Abstract
Deregulation of the EVI1 proto-oncogene by the GATA2 distal hematopoietic enhancer (G2DHE) is a key event in high-risk acute myeloid leukemia carrying 3q21q26 aberrations (3q-AML). Upon chromosomal rearrangement, G2DHE acquires characteristics of a super-enhancer and causes overexpression of EVI1 at 3q26.2. However, the transcription factor (TF) complex of G2DHE remains poorly characterized. The aim of this study was to unravel key components of G2DHE-bound TFs involved in the deregulation of EVI1. We have identified several CEBPA and RUNX1 binding sites to be enriched and critical for G2DHE function in 3q-AML cells. Using ChIP-SICAP (ChIP followed by selective isolation of chromatin-associated proteins), a panel of chromatin interactors of RUNX1 and CEBPA were detected in 3q-AML, including PARP1 and IKZF1. PARP1 inhibition (PARPi) caused a reduction of EVI1 expression and a decrease in EVI1–G2DHE interaction frequency, highlighting the involvement of PARP1 in oncogenic super-enhancer formation. Furthermore, 3q-AML cells were highly sensitive to PARPi and displayed morphological changes with higher rates of differentiation and apoptosis as well as depletion of CD34 + cells. In summary, integrative analysis of the 3q-AML super-enhancer complex identified CEBPA and RUNX1 associated proteins and nominated PARP1 as a potential new therapeutic target in EVI1 + 3q-AML.
Similar content being viewed by others
Introduction
Aberrant expression of the zinc finger transcription factor (TF) Ecotropic Viral Integration Site 1 (EVI1) is a potent oncogenic event involved in the pathogenesis of high-risk hematopoietic neoplasms [1, 2]. EVI1 is transcribed from the MECOM locus on chromosome 3q26.2, which also encodes the Myelodysplasia Syndrome-associated Protein 1 (MDS1) and the longer splice form MDS1-EVI1 [3, 4]. EVI1 and its isoforms are part of the PRDI-BF1 (positive regulatory domain I-binding factor 1) and RIZ1 (retinoblastoma protein-interacting zinc finger gene 1) homology domain (PRDM) family of epigenetic remodelers, with EVI1 also being known as PRDM3 [5, 6]. Although its exact molecular function is still poorly understood, EVI1 plays an essential role in a range of transcriptional regulatory networks important for stem cell maintenance and chromatin remodeling [7,8,9]. EVI1 is known as the most oncogenic isoform and widely studied in leukemia, while MDS1-EVI1 is thought to function as a tumor suppressor [10, 11]. Imbalanced expression of MECOM-transcribed isoforms is often the consequence of (i) chromosomal rearrangements leading either to EVI1 gene fusions (e.g., AML1-EVI1) [12]; (ii) MECOM locus amplifications as found with high frequency in high-grade serous ovarian cancer [13]; (iii) aberrant promoter activation [14]; or (iv) displacement of regulatory DNA elements into the EVI1 locus, the latter being a hallmark of 3q26.2/MECOM rearranged acute myeloid leukemia (AML) [15, 16].
In AML, 3q26.2 rearrangements targeting the MECOM locus typically abrogate expression of both the long MDS1-EVI1 isoform and, coincidentally, expression of other key myeloid regulators, such as GATA2 or MYC, while transcription of EVI1 becomes excessively increased [15,16,17,18]. Similar to EVI1 overexpression caused by inv(3)/t(3;3) in AML, also other PRDM family members are found upregulated in leukemias as for example PRDM16 (MEL1) in t(1;3)/MEL1 rearranged AML [19]. The underlying mechanism of forced EVI1 or MEL1 transcription and co-silencing of GATA2 in 3q-rearranged AML is the repositioning of a GATA2 distal hematopoietic enhancer (G2DHE) into the vicinity of these EVI1 homologs [16, 18, 20]. Relocation of non-coding regulatory DNA elements has recently been coined as enhancer hijacking, and we and others have previously shown that the specific G2DHE element in 3q-rearranged AML is a monoallelic super-enhancer formed on the oncogenic EVI1 allele [15,16,17, 21].
To date, little is known about the molecular composition of the TFs occupying this oncogenic super-enhancer that regulates EVI1, obscuring the identification of novel therapeutic targets that are needed to treat this almost invariably fatal type of AML [22]. In order to better understand the regulation of EVI1 in 3q-rearranged AML, we have performed proteomic analyses of chromatin-bound TFs known to be present at G2DHE, such as CEBPA and RUNX1, and have identified colocalizing complex members of the super-enhancer. Among these, we have focused on translational targets for further characterization in vitro as these may have potential therapeutic significance.
Methods
PARP1 inhibitors
Olaparib (AZD2281; Selleckchem, Houston, TX, USA) and talazoparib (T6253; TargetMol, Wellesley Hills, MA, USA) were resuspended in DMSO at a concentration of 100 mM or 200 mM, respectively.
Dual-luciferase reporter assay
The pGL3 basic vector (Promega, Madison, WI, USA) and the pRL-SV40 Renilla vector (Promega, Madison, WI, USA) were used in all luciferase assays. The pGL3 vector containing the EVI1 promoter was a gift from K. Mitani, Dokkyo Medical University School of Medicine, Kitakobayashi, Japan. Different G2DHE variants were cloned into this vector using the restriction enzymes BamHI and SalI (Table S1). The pGL3 vector containing the EVI1 promoter and full-length G2DHE was described previously [16]. Site-directed mutagenesis was carried out using the QuikChangeII XL kit (Agilent Technologies, Santa Clara, CA, USA) (Table S2). Mutated constructs were re-cloned into a fresh backbone vector. Cells were seeded at a density of 0.5 × 106 cells/mL and transiently transfected with X-tremeGENE HP DNA (Roche, Basel, Switzerland) transfection reagent according to the manufacturer’s protocol. Per 0.5 × 106 cells 100 ng of pRL-SV40 and 900 ng of the full-length G2DHE constructs were used. Equimolar amounts were used for shorter pGL3 constructs. Cells were harvested 48 h after transfection. Dual-luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol on a Victor X3 plate reader (Perkin Elmer, Waltham, MA, USA). Luciferase signal was normalized to Renilla signal.
In silico prediction of TF binding sites
The sequence of the G2DHE core was fed into the online tools JASPAR2016 (JASPAR database, http://jaspar2016.genereg.net/cgi-bin/jaspar_db.pl?rm=browse&db=core&tax_group=vertebrates) and Alggen Promo (TRANSFAC database version 8.3, http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) [23,24,25]. Matrices for selection of important myeloid TFs were chosen and the sequence was scanned with a threshold of 80% relative profile score for JASPAR2016 or 15% dissimilarity score for Alggen Promo. Predicted TF binding sites (TFBS) were checked for conservation between species. Mutations were aimed to have a relative similarity score of less than 70% with an ideal value below 50–60% (JASPAR2016) or more than 15% dissimilarity score (Alggen Promo).
Lentiviral constructs
Short hairpin RNA (shRNA) knockdown experiments were carried out with the optimized microRNA-30 backbone element (miR-E) for CEBPA, RUNX1 and PARP1 [26]. The SGEP vector was a gift from J. Zuber (Research Institute of Molecular Pathology, Vienna, Austria; Addgene plasmid #111170). Genome-wide sensor-based shRNA predictions were used for the choice of the target sequence [26]. 97-mer Ultramer DNA Oligos (Table S3) were ordered from Integrated DNA Technologies and cloned into the SGEP vector as described previously (Table S1) [26].
For EVI1 knockdown, pLKO.1-Puro (Bob Weinberg, Addgene plasmid #8453) harboring shRNAs targeting EVI1 (TRCN0000002532, TRCN0000002531) or a commercial non-targeting control (SHC002, Sigma-Aldrich, St. Louis, MO, USA) were used.
The EVI1 ORF was cloned into pLenti-CMV-Puro-DEST (w118-1) (Eric Campeau & Paul Kaufman, Addgene plasmid #17452). pLenti-CMV-Puro containing the coding sequence for 1xFlag was used as empty vector control.
A detailed transduction protocol is included in the supplemental methods.
ChIP-SICAP
Chromatin immunoprecipitation with selective isolation of chromatin-associated proteins (ChIP-SICAP) was conducted as previously described with few modifications described in the supplement [27, 28]. In brief, cells fixed with formaldehyde were lysed and the nuclear fraction was extracted. Chromatin was sheared using sonication and incubated with antibodies directed against the respective bait proteins or an IgG control. Protein complexes were purified using magnetic beads. Following immunoprecipitation, the DNA bound to the protein complexes was biotinylated and chromatin-associated complexes were purified using streptavidin beads. Eluted proteins and DNA were used for mass spectrometric analysis and qPCR, respectively.
DNA streptavidin pull-down
Nuclear lysate was incubated with PCR-generated (Table S1), biotinylated DNA probes. DNA probes and associated proteins were purified using streptavidin magnetic beads (S1420S; New England Biolabs, Frankfurt am Main, Germany) and analyzed by SDS-PAGE and western blot. 30 µg of nuclear lysate and 30 µL of the last washing step were used as controls. A detailed protocol is included in the supplemental methods.
Flow cytometry
Apoptosis staining was conducted using the FITC Annexin V Apoptosis Detection Kit I (BD Bioscience, San Jose, CA, USA). Staining for differentiation markers was performed using the following antibodies directed against cell surface markers: CD34−PerCPCy5.5 (#343611; Biolegend, San Diego, CA, USA), CD11b−APC (#301309; BioLegend, San Diego, CA, USA), CD14−APCH7 (MϕP9; BD Biosciences, San Jose, CA, USA). For intracellular staining, either one of the following antibodies were used: cMPO-FITC (sc-51741 FITC; Santa Cruz Biotechnology, Dallas, TX, USA) or γH2AX-Alexa488 (560445, BD Biosciences, San Jose, CA, USA). A detailed staining protocol is attached in the Supplementary methods.
Results
The hijacked G2DHE harbors two conserved sequence modules enriched for RUNX1, CEBPA, and MYB motifs
Hijacking of G2DHE is the underlying molecular event in the pathogenesis of inv(3)/t(3;3) AML, as we and others have previously described [15, 16]. We aimed to better understand the molecular basis of the enhancer function in this AML subset by studying the complex of its associated TFs. The core p300-binding portion of G2DHE consists of a highly conserved bimodular sequence structure (Fig. 1a). TFBS prediction analyses showed predominant enrichment of MYB, RUNX1, and CEBPA motifs in the left (centromeric) part, while other prominent myeloid TFs, such as PU.1, GATA2, TAL1, and IKZF1 motifs clustered in the right (telomeric) region. In EVI1-promoter-luciferase reporter studies, a 755 bp region of the core G2DHE was sufficient to induce reporter gene activity, whereas neither left and right module alone exhibited transactivating potential in the t(3;3) HNT-34 cells (Fig. 1b). In order to assess the importance of individual TFBS in vitro, we generated inactivating mutants of the identified TFBS and studied their impact on reporter gene induction (Figs. 1a and S1a). The majority of mutations (16/21) led to reduced enhancer reporter activity in HNT-34 (Fig. 1c), and integration of data from three inv(3)/t(3;3) cell lines (MOLM-1, MUTZ-3, HNT-34) showed consistent downregulation when TFBS of CEBPA, FLI1, MYB, and RUNX1 were mutated (Fig. S1b). For validation of candidate TFs used in further proteomic studies, we chose RUNX1 and CEBPA, since (i) both TFs are pioneer TFs and key myeloid transcriptional regulators expressed at sufficiently high levels in inv(3)/t(3;3) AML allowing for subsequent protein capture experiments (Fig. S2a, b); (ii) RUNX1 has been implicated in the regulation of EVI1 previously [29]; and (iii) CEBPA mutations are mutually exclusive with inv(3)/t(3;3) in AML [1, 30]. Furthermore, EVI1 expression was downregulated following global miR-E -mediated knockdown of CEBPA and RUNX1 in inv(3) AML cells (Fig. 1d), suggesting functional importance of these myeloid TFs in maintaining EVI1 expression by either direct or indirect effects.
a Schematic overview of the transcription factor binding sites identified with the JASPAR2016 and TRANSFAC databases as well as the respective mutations and truncated enhancer fragments in relation to the previously described p300 chromatin binding peaks in MUTZ-3 and MOLM-1 cells and the conservation across species (SiPhy rate 10 mer) [16, 54]. b, c Luciferase reporter assay in HNT-34. Cells were co-transfected with the luciferase reporter plasmid pGL3 and the Renilla control plasmid. pGL3 empty vector (pGL3), pGL3 vector containing the EVI1 Promoter (Prom), and pGL3 vector containing the EVI1 promoter and G2DHE were used as controls (CTRL). Luciferase signal was normalized to Renilla signal. The relative luciferase signal was further normalized to the signal of full-length, wildtype G2DHE (red line). Statistical significance was calculated using two-sided one-sample t-tests. b shows the activity of the truncated enhancer fragments (n = 4) and c shows the activity of enhancer mutants (n ≥ 3). TFBS in parentheses are in close proximity to the mutations without being affected by them. d Protein expression quantification following CEBPA and RUNX1 knockdown in MUTZ-3 cells using miR-E constructs (n = 2 for REN, n = 3 for all other samples). Cells were lentivirally transduced and selected with puromycin. Samples were harvested on day 3 of puromycin selection, and protein levels were analyzed by western blot using antibodies against CEBPA, RUNX1, and EVI1. Western blot signal of the proteins of interest was normalized to the respective loading control and to the scrambled control (SCR). A miR-E construct targeting Renilla (REN) and untreated parental cells serve as additional non-targeting controls. Statistical significance was calculated using two-sided one-sample t-tests.
Enhancer-bound CEBPA and RUNX1 colocalize with IKZF1 and PARP1
To identify proteins that colocalize with CEBPA and RUNX1 in enhancer-bound TF complexes and that are potential therapeutic targets in 3q-rearranged AML, we performed ChIP-SICAP, a method that allows for the isolation of protein complexes in their chromatin-bound state by using specific bait proteins [27]. CEBPA and RUNX1 served as baits, and their locus-specific association with G2DHE was confirmed by ChIP-Seq (Fig. S2c) and qPCR on DNA purified after ChIP-SICAP in the three inv(3)/t(3;3) AML cell lines MUTZ-3, HNT-34, and MOLM-1 (Figs. 2a and S2d). Next, the protein fractions of genomic loci captured with the bait proteins were analyzed by quantitative mass spectrometry (Figs. 2b, S2e–f, and File S1). Successful enrichment of chromatin-associated proteins was evidenced by the detection of histones and by nuclear localization of identified proteins of at least 76% of all captured proteins, as well as by chromatin binding and transcription as some of the most significantly enriched biological processes and molecular functions in all cell lines using both baits according to Gene Ontology (GO) analysis (Figs. 2b and 3a, b). Following this analysis, the majority of all identified proteins were categorized as potential true positives (Fig. S3c). We detected 46 (MUTZ-3), 44 (MOLM-1), and 87 (HNT-34) proteins when using CEBPA as bait, and 108 (MUTZ-3), 74 (MOLM-1), and 151 (HNT-34) proteins following RUNX1 capture (Fig. 2c). Already known interactors of the bait proteins CEBPA (17% of all identified ChIP-SICAP proteins) and RUNX1 (33% of all identified ChIP-SICAP proteins) were detected using Biological General Repository for Interaction Datasets (BioGRID) analyses (Fig. S3a, b and Table S4). When comparing different cell lines, 30 proteins were identified in at least two cell lines using CEBPA as bait, and 51 for RUNX1 (Fig. 2d). Within the CEBPA and RUNX1 datasets, we found a total of 19 proteins to be enriched for both bait proteins in at least two inv(3)/t(3;3) cell lines (Fig. 2d). These included previously described enhancer-associated proteins, such as the histone acetyltransferase p300 (EP300) that initially led us to the identification of the G2DHE [16]. Furthermore, among the recurrently detected candidates were Ikaros family zinc finger 1 (IKZF1), a TF previously described in inv(3)/t(3;3) AML, and poly(ADP-ribose) polymerase 1 (PARP1) (Fig. 2c, d) [31].
ChIP-SICAP was performed in three different 3q-rearranged cell lines using antibodies against CEBPA and RUNX1 or an unspecific IgG control. Experiments were carried out in duplicates (n = 2). a, b show exemplary results for the cell line MUTZ-3. a The DNA fraction of the ChIP-SICAP experiment was used for enrichment quantification of the G2DHE region by qPCR compared to two unrelated control regions (CTRL). b The protein fraction of the ChIP-SICAP experiments was analyzed by mass spectrometry to identify chromatin-bound interactors of RUNX1 and CEBPA. Proteins enriched in the CEBPA- or RUNX1-captured samples over the IgG controls in both MUTZ-3 replicates were ranked according to their iBAQ intensity. c Overlap of the proteins identified in different cell lines. d Overlap of proteins that were identified in at least two cell lines per bait. e DNA-streptavidin pull-down (PD) of CEBPA, RUNX1, IKZF1, and PARP1. Nuclear lysate of MUTZ-3 cells was incubated with biotinylated DNA probes containing the G2DHE sequence or a deserted control region from chromosome Y (ChrY CTRL). Proteins associated with the DNA probes were purified and analyzed by western blot using antibodies against CEBPA, RUNX1, IKZF1, and PARP1. Input and an aliquot of the last wash steps (W) served as control for the PD samples. f Co-immunoprecipitation of PARP1 and RUNX1. Total cell lysate of MUTZ-3 was used for immunoprecipitation either with a PARP1 antibody or a control IgG. 75 µg of lysate was used as input control. Western blot was performed using antibodies directed against PARP1 and RUNX1.
a Correlation of EVI1 and PARP1 normalized transcripts per million (TPM) expression levels. Values were measured by RNA-Seq in human primary AML patient samples (n = 73) and cell lines (n = 9) as described previously [16]. Shown is the Pearson correlation coefficient (R) with p-value (p). b EVI1 mRNA expression after PARPi. Cells were treated with 10 µM olaparib (ola), 1 µM talazoparib (tala), or DMSO or were left untreated. EVI1 mRNA expression was analyzed by qPCR following PARPi treatment for 24 h (n = 3). Results are normalized to the housekeeping gene HMBS and the DMSO sample. Statistical significance was calculated using two-sided two-sample t-tests. c Normalized read counts of MUTZ-3 treated with DMSO, 10 µM olaparib (ola), or 1 µM talazoparib (tala) across multiple time points as determined by RNA-Seq. d Western blot analysis of EVI1 protein levels in MUTZ-3 cells after 24 h PARPi with 10 µM olaparib (ola) or 1 µM talazoparib (tala). The protein ladder is marked with L. e Protein expression quantification following PARP1 knockdown in MUTZ-3 cells using miR-E constructs (n = 2). Cells were lentivirally transduced and selected with puromycin. Samples were harvested on day 3 of puromycin selection, and protein levels were analyzed by western blot using antibodies against PARP1 and EVI1. Western blot signal of the proteins of interest was normalized to β-Actin and to the parental control. A miR-E construct targeting Renilla (REN) and untreated parental cells served as non-targeting controls. f Semiquantitative analysis of the local chromatin interaction profile of the EVI1 promoter at the 3q21 G2DHE location as determined by 4C-Seq in MUTZ-3 cells following treatment with 10 µM olaparib, 1 µM talazoparib, or DMSO for 24 h. The 4 C signal is measured by calculation of a sliding window average (running mean) of the normalized read counts (window size: 21 fragment ends). The 4 C tracks were group autoscaled and the numbers indicate the data range.
Since ChIP-SICAP represents a global interrogation of interactors with bait-bound chromatin loci genome-wide, we subsequently tested binding of IKZF1 and PARP1 to G2DHE sequence probes in nuclear lysates of MUTZ-3 cells (Fig. 2e). Compared to an unrelated control probe covering a desert chromatin locus on chromosome Y, both CEBPA and RUNX1 bait proteins as well as IKZF1 and PARP1 were detected (Fig. 2e). This confirms that the proteins identified with ChIP-SICAP can associate with the G2DHE sequence in vitro and thus may represent rational targets for further validation. Moreover, interaction of PARP1 and RUNX1 was further validated by co-immunoprecipitation (Fig. 2f). Interaction of CEBPA and PARP1 has been previously demonstrated in prostate cancer, however, we could not validate their interaction in AML cells due to overlap with unspecific antibody/bead complex signals [32].
PARP1 inhibition reduces EVI1 expression
Besides its canonical role in DNA repair, PARP1 has been found to have important functions in cooperating with cellular key identity TFs to organize chromatin structures, bind enhancers, and create chromatin environments permissive of transcription in embryonic stem cell and neural differentiation [33, 34]. We thus hypothesized that PARP1 might be integral to the establishment of G2DHE into an oncogenic super-enhancer on the rearranged 3q allele and necessary to maintain EVI1 expression in inv(3)/t(3;3) AML. EVI1 and PARP1 mRNA expression was significantly correlated as determined by RNA-Seq analysis of previously described primary AML patient samples and cell lines (Fig. 3a and File S2) [16], suggesting coregulation of the two genes in AML. It has recently been shown that PARP1-mediated establishment of activating enhancer-promoter interactions are dependent on the catalytic activity of PARP1 [35]. To assess whether PARP1 activity is required for EVI1 transcription, we treated MUTZ-3 cells with the PARP1 inhibitors olaparib (10 µM) and talazoparib (1 µM) (Fig. 3b), both being potent inhibitors of the catalytic activity of PARP1, while talazoparib also causes trapping of PARP1 on chromatin. For both compounds, we found a significant reduction in EVI1 mRNA expression following PARP1 inhibition (PARPi) as compared to vehicle control after 24 h. RNA-Seq confirmed the decrease of EVI1 expression in a time-dependent manner upon PARPi (Fig. 3c). This effect was evident even more strongly on protein level (Figs. 3d and S4a). Additionally, similar effects could be observed upon shRNA-mediated PARP1 knockdown (Figs. 3e and S4b). In order to determine if EVI1 downregulation was accompanied by reduced chromatin interaction between the EVI1 promoter and the rearranged G2DHE upon PARPi, we performed 4C-Seq in MUTZ-3 cells after 24 h of PARPi treatment. Using the EVI1 promoter as viewpoint, 4C-Seq comparative analysis revealed a decreased interaction frequency between the rearranged G2DHE and the EVI1 promoter in response to PARPi, corroborating previous reports on the necessity of PARP1 occupancy of certain enhancers in long-range gene promoter interactions (Fig. 3f) [35,36,37].
PARP1 inhibition affects cell survival and morphology
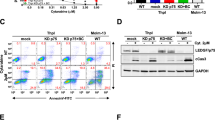
Olaparib and talazoparib treatment of MUTZ-3 cells led to a substantial reduction in cell viability (Fig. 4a). The growth inhibitory effect of PARPi was also apparent in various other 3q-rearranged AML cells lines MOLM-1, HNT-34, and UCSD-AML1 in a concentration- and time-dependent manner (Figs. 4b and S5) with higher efficacy of talazoparib compared to olaparib, while the non-3q-rearranged AML cell lines U-937 and K-562 showed a tendency to be less sensitive towards the PARPi treatment. When treating MUTZ-3 cells with 1 µM talazoparib or 10 µM olaparib, the reduction of cell viability of AML cells went along with a higher percentage of apoptotic cells as shown by Annexin-V/7-AAD staining in flow cytometric analysis (Fig. 4c). Similar to effects previously seen upon EVI1 downregulation induced by G2DHE deletion, immunophenotyping of PARPi treated inv(3) MUTZ-3 cells showed a loss of CD34 expression along with an increase in myelomonocytic differentiation as measured by CD14 expression (intermediate: CD34−/CD14− and mature: CD34−, CD14+), whereas the immature progenitor cells (CD34+/CD14−) were almost completely lost in comparison to the controls (Fig. 4d). Furthermore, cell morphology assessment of PARPi treated cells using May–Grünwald–Giemsa staining confirmed morphological changes with bigger cytoplasm, more vacuolization, and appearance of monocyte-like cells after treatment with talazoparib or olaparib compared to the DMSO vehicle control and untreated cells after 24 h (Fig. 4e).
a, b Sensitivity of different cell lines to PARPi. Cells were treated with the indicated amounts of olaparib or talazoparib. Shown is the mean ± SD (n = 3). Metabolic activity was measured as an indicator of cell viability by CellTiter-Glo assay. Values were normalized to those of the 0 h time point and to the DMSO control of each time point. a Viability of MUTZ-3 cells across multiple time points. b Comparison of several 3q-rearranged and non-rearranged cell lines at 72 h. c–e Effects of PARPi treatment in MUTZ-3 cells. Cells were treated with 10 µM olaparib (ola), 1 µM talazoparib (tala), or DMSO or were left untreated for 24 h. c Apoptosis staining with Annexin-V and 7-AAD and flow cytometric analysis. The graphs show one representative replicate. d Flow cytometric analysis of differentiation markers. Cells were gated for the live population. The graphs show one representative replicate. e Representative images of May-Grünwald-Giemsa staining of control and PARPi treated MUTZ-3 cells. f EVI1 overexpression partially rescues loss of immature cells after PARPi. MUTZ-3 cells were transduced using a lentiviral expression vector containing EVI1 or the corresponding empty vector (EV) control. After selection with puromycin, cells were treated with 10 µM olaparib or DMSO for 24 h. Differentiation markers were assessed by flow cytometry. Cells were gated for the live cell population. The amount of immature (CD34 + /CD14−) cell population of PARPi treated cells was normalized to the DMSO control. The graph shows one representative replicate. g DNA damage response after PARPi in MUTZ-3 and U-937. Intracellular γH2AX levels of untreated cells or cells treated with DMSO, 10 µM olaparib, or 1 µM talazoparib were assessed by flow cytometric analysis. Cells were gated for the single cell population. h Commonly downregulated (left) and upregulated (right) genes by PARPi and EVI1 knockdown as identified by RNA-Seq. Genes included in cluster 3 of the PARPi data were compared to the genes deregulated by EVI1 knockdown to determine the genes downregulated by both conditions. Upregulated genes in cluster 4 of the PARPi RNA-Seq data were determined accordingly. Fold change of gene expression under treatment conditions (PARPi or shEVI1, respectively) over the control (DMSO or non-targeting control, respectively) is shown. Exemplary genes are annotated.
Next, we investigated whether the phenotype observed after PARPi was primarily dependent on epigenetic downregulation of EVI1 transcription or rather secondary to PARP1-specific functional effects, such as induction of DNA damage. Forced expression of EVI1 partially rescued the loss of immature progenitor cells upon PARPi treatment (Fig. 4f), suggesting a direct interaction between EVI1 and PARP1. Furthermore, DNA damage was monitored by assessing intracellular γH2AX levels (Fig. 4g). PARPi induced a large increase of γH2AX in the 3q-negative cell line U-937, however, in the 3q-rearranged cell line MUTZ-3 only a low to moderate increase was observable.
RNA-Seq analysis of PARPi treated MUTZ-3 cells revealed 4 distinct clusters among the 1 500 most differentially regulated genes (Fig. S6a and File S3). Of those, cluster 2 (159 genes) and cluster 4 (756 genes) showed higher gene expression after PARPi, whereas cluster 3 (454 genes) was downregulated. Pathway enrichment analysis of the different clusters (Fig. S6b–e) identified cell cycle control and DNA repair-related pathways to be enriched among genes downregulated after PARPi treatment (Fig. S6d). Neutrophil degranulation, interferon signaling, and other immune system-related pathways were enriched among upregulated genes (Fig. S6e), which supports the observation of a differentiation phenotype of inv(3) AML cells following PARPi treatment (Fig. 4d–e). Correlation of RNA-Seq data upon either PARPi (cluster 3) or EVI1 knockdown (File S3) revealed that 46 genes were commonly downregulated between the two conditions, including myeloid regulators CD34, ERG, HOXB2, and EVI1 itself (Figs. 4h and S7a, b). 91 genes were found to be commonly upregulated between the two conditions when using PARPi cluster 4 for comparison with the EVI1 knockdown (Figs. 4h and S7c, d), including genes involved in immune cell maturation like CD14.
Overall, these data indicate that DNA damage and impaired DNA damage response do not appear to be the primary cause of PARPi sensitivity and subsequent EVI1 transcriptional disturbance in the 3q-rearranged MUTZ-3 cells.
Discussion
The context-dependent oncogenic functions of EVI1 family members have increasingly been recognized in multiple types of cancer, although the mechanisms of oncogenic transformation caused by EVI1 are complex and incompletely understood [11, 13, 38]. Previous reports support the notion that the tight interplay between deregulated EVI1 and simultaneous disturbances in the dosage of other myeloid TFs, such as GATA2, is linked to the development and maintenance of therapy-resistant AML with 3q26.2/EVI1 rearrangements [16,17,18, 20]. From these studies, G2DHE emerged as a common regulatory node between these TFs and as a key regulator of hematopoiesis with leukemogenic consequences when misplaced [15,16,17,18, 20, 29]. However, the molecular architecture and functional characteristics of G2DHE have been little understood, particularly with respect to EVI1-rearranged AML. This prompted us to investigate the TF composition of G2DHE and potential interactors.
Our studies provide molecular and functional evidence of the TF composition of G2DHE in inv(3)/t(3;3) AML that may render it a therapeutic liability for this disease category. The interplay and additive effect of those TFBS is integral to G2DHE function as both modules are required for maximum enhancer activity. Among the most recurrent and functionally important TFs in reporter studies of G2DHE in vitro were RUNX1, MYB, CEBPA, IKZF1, FLI1, ELK1, and GATA2 itself, which play key roles in hematopoiesis [39,40,41]. In line with our observations, RUNX1, MYB, and ELK1 have previously been implicated in the regulation of EVI1 transcription in AML cells by studies of the minimal EVI1 promoter of the MECOM locus, which is the target of the hijacked G2DHE in 3q-rearranged AML [29]. Although, CEBPA has been reported to be negatively regulated by EVI1, we have observed measurably high expression of CEBPA in inv(3)/t(3;3) cell lines (Fig. S2b), clear binding of CEBPA at G2DHE in inv(3)/t(3;3) cells, and activating potential in reporter experiments [42]. The presence of RUNX1 and CEBPA binding at G2DHE allowed targeted chromatin-capture and thereby identification of novel interacting partners. A limitation of ChIP-SICAP is its lack of targeted identification of locus-specific interactions, thus it was not possible to discriminate between TF complexes of the orthotopic and rearranged G2DHE alleles. However, histone ChIP-Seq data point to the generation of a super-enhancer accessible for TFs only on the rearranged 3q21 allele [16].
ChIP-SICAP was used to identify a panel of CEBPA and RUNX1 interactors. Association of the two hits PARP1 and IKZF1 was confirmed by an in vitro pull-down approach (Fig. 2e); however, ChIP(-Seq) of these two hits in our 3q-rearranged model cell lines proved not to be feasible.
The mapping of IKZF1 DNA binding sites in G2DHE impacting EVI1 promoter activity and chromatin co-occupancy with RUNX1 and CEBPA as found in ChIP-SICAP experiments implicate an interaction of IKZF1 with EVI1/GATA2 pathways, which was previously indicated by a study investigating the mutational landscape of EVI1-rearranged AML [31]. We have observed a slight impact on EVI1 expression, but no reduced viability of inv(3)/t(3;3) AML cell lines in response to lenalidomide treatment (data not shown), which causes selective degradation of IKZF1 [43]. Notwithstanding, the use of lenalidomide has been suggested as a possible adjunct to hypomethylating agents in the treatment of inv(3)/t(3;3) AML patients [44].
PARP1 was identified in ChIP-SICAP experiments using both G2DHE baits RUNX1 and CEBPA in different 3q-rearranged cell lines. Inhibition of PARP1 catalytic activity and PARP1 knockdown led to downregulation of EVI1. RNA-Seq analysis showed that more genes were upregulated than downregulated after PARPi treatment (Fig. S6a). Hence, it appears that instead of causing a global decrease of transcription, the downregulation of EVI1 does not appear to be unspecific. Further analysis of cells treated with PARPi also showed a profound growth arrest of inv(3) AML cells, and increased differentiation, which can be partially rescued by overexpression of EVI1. This differentiation pattern was similar to that observed after EVI1 knockdown, BRD4 inhibition, or G2DHE deletion previously [16]. Comparative RNA-Seq analysis following PARPi and EVI1 knockdown confirmed induction of differentiation gene expression patterns and revealed a subset of coregulated genes. Interestingly, one of the strongest negatively regulated gene both in PARPi and EVI1 knockdown in inv(3) AML cells was PHGDH, a serine biosynthesis enzyme implicated as a critical metabolic regulator necessary for propagation of leukemia cells and other cancers [45,46,47].
Previous studies showed that PARP1 can exert its positive gene regulatory function by poly(ADP-ribosyl)ation (PARylation) of histones and the histone-associated protein DEK (Fig. 2b) and the histone deacetylase SIRT, leading to chromatin decompaction, binding of Mediator coregulatory complex, and initiation of transcription [48,49,50]. Here, PARPi led to reduced G2DHE – EVI1 promoter interaction frequency, indicating that the persistent activity of the oncogenic super-enhancer relies on the constitutive presence and chromatin modulation of PARP1. PARP1-mediated chromatin remodeling has been shown to be a facilitator of binding of the pioneer TF Sox2 to intractable enhancers in closed chromatin regions of embryonic stem cells, making PARP1 a requirement for the expression of pluripotency genes and stemness [37]. Conversely, a recent report indirectly implicated PARP1 in the downregulation of genes involved in immune evasion of leukemic stem cells, since PARPi induced expression of natural killer cell ligands, although the exact mechanism of PARPi-mediated remained obscure [51]. Altogether, these observations confirm the novel molecular function of PARP1 in chromatin regulation, highlight its importance in oncogene regulation in leukemia with enhancer hijacking due to chromosomal rearrangements, and make PARP1 a potential attractive target for further clinical investigation.
References
Gröschel S, Lugthart S, Schlenk RF, Valk PJM, Eiwen K, Goudswaard C, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28:2101–7.
Lugthart S, Van Drunen E, Van Norden Y, Van Hoven A, Erpelinck CAJ, Valk PJM, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: Prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–37.
Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–40.
Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WLJ, Valk PJM, van der Poel-van de Luytgaarde S, Hack R, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–45.
Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene 2006;368:1–11.
Lin Y, Wong KK, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–9.
Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–20.
Ivanochko D, Halabelian L, Henderson E, Savitsky P, Jain H, Marcon E, et al. Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex. Nucleic Acids Res. 2019;47:1225–38.
Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–60.
Sood R, Talwar-Trikha A, Chakrabarti SR, Nucifora G. MDS1/EVI1 enhances TGF-β1 signaling and strengthens its growth-inhibitory effect, but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-β1. Leukemia. 1999;13:348–57.
Hinai AA, Valk PJM. Review: aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172:870–8.
Mitani K, Ogawa S, Tanaka T, Miyoshi H, Kurokawa M, Mano H, et al. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 1994;13:504–10.
Nanjundan M, Nakayama Y, Kwai WC, Lahad J, Liu J, Lu K, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67:3074–84.
Bindels EMJ, Havermans M, Lugthart S, Erpelinck C, Wocjtowicz E, Krivtsov AV, et al. EVI1 is critical for the pathogenesis of a subset of MLL-AF9-rearranged AMLs. Blood. 2012;119:5838–49.
Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25:415–27.
Gröschel S, Sanders MA, Hoogenboezem R, De Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in Leukemia. Cell. 2014;157:369–81.
Ottema S, Mulet-Lazaro R, Beverloo HB, Erpelinck CAJ, van Herk S, van der Helm R, et al. Atypical 3q26/MECOM rearrangements genocopy inv(3)/t(3;3) in acute myeloid leukemia. Blood. 2020;136:224–34.
Yamaoka A, Suzuki M, Katayama S, Orihara D, Engel JD, Yamamoto M. EVI1 and GATA2 misexpression induced by inv(3)(q21q26) contribute to megakaryocyte-lineage skewing and leukemogenesis. Blood Adv. 2020;4:1722–36.
Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–14.
Katayama S, Suzuki M, Yamaoka A, Keleku-Lukwete N, Katsuoka F, Otsuki A, et al. GATA2 haploinsufficiency accelerates EVI1-driven leukemogenesis. Blood. 2017;130:908–19.
Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–34.
Lugthart S, Gröschel S, Beverloo HB, Kayser S, Valk PJM, Van Zelderen-Bhola SL, et al. Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28:3890–8.
Messeguer X, Escudero R, Farré D, Nuñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4.
Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3.
Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115.
Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13.
Rafiee MR, Girardot C, Sigismondo G, Krijgsveld J. Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol Cell. 2016;64:624–35.
Rafiee M, Sigismondo G, Kalxdorf M, Förster L, Brügger B, Béthune J, et al. Protease-resistant streptavidin for interaction proteomics. Mol Syst Biol. 2020;16:e9370.
Maicas M, Vazquez I, Vicente C, Garcıa-Sanchez MA, Marcotegui N, Urquiza L, et al. Functional characterization of the promoter region of the human EVI1 gene in acute myeloid leukemia: RUNX1 and ELK1 directly regulate its transcription. Oncogene. 2013;32:2069–78.
Gröschel S, Sanders MA, Hoogenboezem R, Zeilemaker A, Havermans M, Erpelinck C, et al. Mutational spectrum of myeloid malignancies with inv(3)/t(3;3) reveals a predominant involvement of RAS/RTK signaling pathways. Blood. 2015;125:133–9.
Lavallée VP, Gendron P, Lemieux S, D’Angelo G, Hébert J, Sauvageau G. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood. 2015;125:140–3.
Yin H, Glass J. In prostate cancer cells the interaction of C/EBPalpha with Ku70, Ku80, and poly(ADP-ribose) polymerase-1 increases sensitivity to DNA damage. | BioGRID. J Biol Chem. 2006;281:11496–505.
Iglehart JD, Silver DP. Synthetic lethality - A new direction in cancer-drug development. N. Engl J Med 2009;361:189–91.
Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24.
Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, et al. Decreased enhancer-promoter proximity accompanying enhancer activation. Mol Cell. 2019;76:473–84.
Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–21.
Liu Z, Kraus WL. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol Cell. 2017;65:589–603.
Wang H, Schaefer T, Konantz M, Braun M, Varga Z, Paczulla AM, et al. Prominent oncogenic roles of EVI1 in breast carcinoma. Cancer Res. 2017;77:2148–60.
Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–6.
Gao X, Johnson KD, Chang YI, Boyer ME, Dewey CN, Zhang J, et al. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–42.
Wlodarski MW, Collin M, Horwitz MS. GATA2 deficiency and related myeloid neoplasms. Semin Hematol. 2017;54:81–86.
Wilson M, Tsakraklides V, Tran M, Xiao YY, Zhang Y, Perkins AS. EVI1 interferes with myeloid maturation via transcriptional repression of Cebpa, via binding to two far downstream regulatory elements. J Biol Chem. 2016;291:13591–607.
Lu G, Middleton RE, Sun H, Naniong MV, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science. 2014;343:305–9.
Merz LE, Perissinotti AJ, Marini BL, Burke PW, Crouch A, Erba HP, et al. Lenalidomide plus hypomethylating agent as a treatment option in acute myeloid leukemia with recurrent genetic abnormalities—AML with inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2, MECOM. Clin Lymphoma, Myeloma Leuk. 2020;20:24–30.
Kampen KR, Fancello L, Girardi T, Rinaldi G, Planque M, Sulima SO, et al. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat Commun. 2019;10:2542.
Polet F, Corbet C, Pinto A, Rubio LI, Martherus R, Bol V, et al. Reducing the serine availability complements the inhibition of the glutamine metabolism to block leukemia cell growth. Oncotarget. 2016;7:1765–76.
Sharif T, Martell E, Dai C, Ghassemi-Rad MS, Lee K, Singh SK, et al. Phosphoglycerate dehydrogenase inhibition induces p-mTOR-independent autophagy and promotes multilineage differentiation in embryonal carcinoma stem-like cells. Cell Death Dis. 2018;9:990.
Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–55.
Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–67.
Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-Ribose) polymerase promotes nucleosome Loss at Hsp70. Mol Cell. 2012;45:64–74.
Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang H, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572:254–9.
Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450.
Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–62.
Acknowledgements
This research was supported with a Starting Grant (677209) of the European Research Council (S.G.), the program “ENHancers And Non-coding Cancer (Epi-)Mutations (ENHANCE)” funded within the DKFZ-NCT3.0 initiative on “Integrative Projects in Basic Cancer Research” (CH, SG); a scholarship of the Helmholtz International Graduate School for Cancer Research Graduate School (JMi, SKi); and the Excellence Cluster CellNetworks (JK). SKi is a PhD candidate at the University of Heidelberg and the DKFZ. This work is submitted in partial fulfillment of the requirement for the PhD. We thank Claudia Scholl and Stefan Fröhling (German Cancer Research Center, Heidelberg, Germany and Nation Center for Tumor Diseases, Heidelberg, Germany), Marie Groth (German Cancer Research Center, Heidelberg, Germany and Novartis, Nürnberg, Germany) as well as Carolin Andresen and Daniel Hübschmann (German Cancer Research Center, Heidelberg, Germany and Heidelberg Institute for Stem cell Technology and Experimental Medicine, Heidelberg, Germany) for their support. We thank the DKFZ Genomics and Proteomics Core Facility, Tim Kühnenz and Mathias Utz (German Cancer Research Center, Heidelberg, Germany) for technical assistance.
Data sharing and declaration
The ChIP-SICAP mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [52] partner repository with the dataset identifiers PXD020045, PXD020050, and PXD020054. ChIP-Seq, RNA-Seq, and 4C-Seq data have been deposited at NCBI’s Gene Expression Omnibus [53] and are accessible through GEO series accession number GSE153307.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
SKi, JMi, JMü, SG, SS, GS, and AB performed experiments; SKi, M-RR, SKr, CH, and AB analyzed results; SKi made the figures; SKi, M-R, PS, JK, and SG designed the experiments; SKi and SG wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kiehlmeier, S., Rafiee, MR., Bakr, A. et al. Identification of therapeutic targets of the hijacked super-enhancer complex in EVI1-rearranged leukemia. Leukemia 35, 3127–3138 (2021). https://doi.org/10.1038/s41375-021-01235-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01235-z
This article is cited by
-
High EVI1 and PARP1 expression as favourable prognostic markers in high-grade serous ovarian carcinoma
Journal of Ovarian Research (2023)
-
TET2 lesions enhance the aggressiveness of CEBPA-mutant acute myeloid leukemia by rebalancing GATA2 expression
Nature Communications (2023)