Abstract
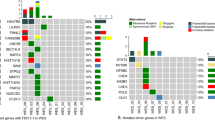
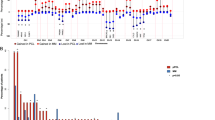
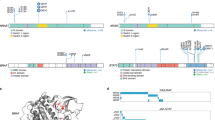
POEMS syndrome is a rare paraneoplastic disease associated with monoclonal plasma cells; however, the pathogenic importance of plasma cells remains unclear. We performed comprehensive genetic analyses of plasma cells in 20 patients with POEMS syndrome. Whole exome sequencing was performed in 11 cases and found a total of 308 somatic mutations in 285 genes. Targeted sequencing was performed in all 20 cases and identified 20 mutations in 7 recurrently mutated genes, namely KLHL6, LTB, EHD1, EML4, HEPHL1, HIPK1, and PCDH10. None of the driver gene mutations frequently found in multiple myeloma (MM) such as NRAS, KRAS, BRAF, and TP53 was detected. Copy number analysis showed chromosomal abnormalities shared with monoclonal gammopathy of undetermined significance (MGUS), suggesting a partial overlap in the early development of MGUS and POEMS syndrome. RNA sequencing revealed a transcription profile specific to POEMS syndrome when compared with normal plasma cells, MGUS and MM. Unexpectedly, disease-specific VEGFA expression was not increased in POEMS syndrome. Our study illustrates that the genetic and transcriptional profiles of plasma cells in POEMS syndrome are distinct from MM and MGUS, indicating unique function of clonal plasma cells in its pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Takatsuki K, Sanada I. Plasma cell dyscrasia with polyneuropathy and endocrine disorder: clinical and laboratory features of 109 reported cases. Jpn J Clin Oncol. 1983;13:543–55.
Nakanishi T, Sobue I, Toyokura Y, Nishitani H, Kuroiwa Y, Satoyoshi E, et al. The Crow-Fukase syndrome: a study of 102 cases in Japan. Neurology. 1984;34:712–20.
Nasu S, Misawa S, Sekiguchi Y, Shibuya K, Kanai K, Fujimaki Y, et al. Different neurological and physiological profiles in POEMS syndrome and chronic inflammatory demyelinating polyneuropathy. J Neurol Neurosurg Psychiatry. 2012;83:476–9.
Bardwick PA, Zvaifler NJ, Gill GN, Newman D, Greenway GD, Resnick DL. Plasma cell dyscrasia with polyneuropathy, organomegaly, endocrinopathy, M protein, and skin changes: the POEMS syndrome. Report on two cases and a review of the literature. Medicine. 1980;59:311–22.
Dispenzieri A. POEMS syndrome: update on diagnosis, risk-stratification, and management. Am J Hematol. 2015;90:951–62.
Misawa S, Sato Y, Katayama K, Hanaoka H, Sawai S, Beppu M, et al. Vascular endothelial growth factor as a predictive marker for POEMS syndrome treatment response: retrospective cohort study. BMJ Open. 2015;5:e009157.
Kawajiri-Manako C, Sakaida E, Ohwada C, Miyamoto T, Azuma T, Taguchi J, et al. Efficacy and long-term outcomes of autologous stem cell transplantation in POEMS syndrome: a nationwide survey in Japan. Biol Marrow Transplant. 2018;24:1180–6.
Ohwada C, Sakaida E, Kawajiri-Manako C, Nagao Y, Oshima-Hasegawa N, Togasaki E, et al. Long-term evaluation of physical improvement and survival of autologous stem cell transplantation in POEMS syndrome. Blood. 2018;131:2173–6.
Abe D, Nakaseko C, Takeuchi M, Tanaka H, Ohwada C, Sakaida E, et al. Restrictive usage of monoclonal immunoglobulin lambda light chain germline in POEMS syndrome. Blood. 2008;112:836–9.
Martin S, Labauge P, Jouanel P, Viallard JL, Piette JC, Sauvezie B. Restricted use of Vlambda genes in POEMS syndrome. Haematologica. 2004;89:Ecr02.
Li J, Huang Z, Duan MH, Zhang W, Chen M, Cao XX, et al. Characterization of immunoglobulin lambda light chain variable region (IGLV) gene and its relationship with clinical features in patients with POEMS syndrome. Ann Hematol. 2012;91:1251–5.
Nakayama-Ichiyama S, Yokote T, Hirata Y, Iwaki K, Akioka T, Miyoshi T, et al. Multiple cytokine-producing plasmablastic solitary plasmacytoma of bone with polyneuropathy, organomegaly, endocrinology, monoclonal protein, and skin changes syndrome. J Clin Oncol. 2012;30:e91–e94.
Wang C, Huang XF, Cai QQ, Cao XX, Cai H, Zhou D, et al. Remarkable expression of vascular endothelial growth factor in bone marrow plasma cells of patients with POEMS syndrome. Leuk Res. 2016;50:78–84.
Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21.
Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–8.
Korde N, Kristinsson SY, Landgren O. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM): novel biological insights and development of early treatment strategies. Blood. 2011;117:5573–81.
Kanai K, Sawai S, Sogawa K, Mori M, Misawa S, Shibuya K, et al. Markedly upregulated serum interleukin-12 as a novel biomarker in POEMS syndrome. Neurology. 2012;79:575–82.
Gupta-Rossi N, Storck S, Griebel PJ, Reynaud CA, Weill JC, Dahan A. Specific over-expression of deltex and a new Kelch-like protein in human germinal center B cells. Mol Immunol. 2003;39:791–9.
Kroll J, Shi X, Caprioli A, Liu HH, Waskow C, Lin KM, et al. The BTB-kelch protein KLHL6 is involved in B-lymphocyte antigen receptor signaling and germinal center formation. Mol Cell Biol. 2005;25:8531–40.
Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5.
Ganapathi KA, Jobanputra V, Iwamoto F, Jain P, Chen J, Cascione L, et al. The genetic landscape of dural marginal zone lymphomas. Oncotarget. 2016;7:43052–61.
Mikulasova A, Walker BA, Wardell CP, Boyle EM, Murison A, Kufova Z, et al. Somatic mutation spectrum in monoclonal gammopathy of undetermined significance compared to multiple myeloma. Blood. 2014;124:3346. ASH meeting abstract
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72.
Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132:587–97.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98.
Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013;27:1–17.
Palacios F, Moreno P, Morande P, Abreu C, Correa A, Porro V, et al. High expression of AID and active class switch recombination might account for a more aggressive disease in unmutated CLL patients: link with an activated microenvironment in CLL disease. Blood. 2010;115:4488–96.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101.
Walker BA, Wardell CP, Murison A, Boyle EM, Begum DB, Dahir NM, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6:6997.
Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–44.
Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115:3541–52.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–20.
Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic landscapes of relapsed and refractory diffuse large B-Cell lymphomas. Clin Cancer Res. 2016;22:2290–2300.
Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 2011;21:122–31.
Pollmann M, Parwaresch R, Adam-Klages S, Kruse ML, Buck F, Heidebrecht HJ. Human EML4, a novel member of the EMAP family, is essential for microtubule formation. Exp Cell Res. 2006;312:3241–51.
Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–6.
Chen H, Attieh ZK, Syed BA, Kuo YM, Stevens V, Fuqua BK, et al. Identification of zyklopen, a new member of the vertebrate multicopper ferroxidase family, and characterization in rodents and human cells. J Nutr. 2010;140:1728–35.
Rey C, Soubeyran I, Mahouche I, Pedeboscq S, Bessede A, Ichas F, et al. HIPK1 drives p53 activation to limit colorectal cancer cell growth. Cell Cycle. 2013;12:1879–91.
Li Z, Yang Z, Peng X, Li Y, Liu Q, Chen J. Nuclear factor-kappaB is involved in the protocadherin-10-mediated pro-apoptotic effect in multiple myeloma. Mol Med Rep. 2014;10:832–8.
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82.
Nagy A, Bhaduri A, Shahmarvand N, Shahryari J, Zehnder JL, Warnke RA, et al. Next-generation sequencing of idiopathic multicentric and unicentric Castleman disease and follicular dendritic cell sarcomas. Blood Adv. 2018;2:481–91.
Dao LN, Hanson CA, Dispenzieri A, Morice WG, Kurtin PJ, Hoyer JD. Bone marrow histopathology in POEMS syndrome: a distinctive combination of plasma cell, lymphoid, and myeloid findings in 87 patients. Blood. 2011;117:6438–44.
Kawajiri-Manako C, Mimura N, Fukuyo M, Namba H, Rahmutulla B, Nagao Y, et al. Clonal immunoglobulin lambda light-chain gene rearrangements detected by next generation sequencing in POEMS syndrome. Am J Hematol. 2018;93:1161–68.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69.
Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293–9.
Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373:35–47.
Acknowledgements
We thank Rika Okada, Miki Takahashi, and Ayumi Koga for their technical assistance; and all clinical staff involved in patient management. This study was supported in part by Grants-in-Aid for Scientific Research of the Japan Society for the Promotion of Science KAKENHI #26461397 and 15K09494 as well as by Grants-in-Aid for Scientific Research on Innovative Areas “Stem Cell Aging and Disease” #25115002 from MEXT, Japan. E.S. was a recipient of JSM Research Award from the Japanese Society of Myeloma.
Author contributions
Y.N., N.M., A.I., S.O., and C.N. conceived and designed the research. Y.N., N.M., Y.I., K.K., C.K.M., N.O.H., S.T., S.S., Y.T., C.O., M.T., T.I., S.Mis., E.S., S.Ku., and C.N. managed care for patients, collected BM samples, and analyzed clinical data. Y.N., N.M., J.T., K.Yos., Y.Shiozawa, M.O., K.A., A.S., S.Ko., O.R., Y.H., D.N., Y.I., and K.K. performed experiments and statistical analyses. Y.Shiraishi, K.C., H.T., and S.Miyano developed the study methodology. Y.N., N.M., J.T., K.Yos., M.O., O.O., K.Yok., M.S., A.I., S.O., and C.N. interpreted data. Y.N. and N.M. actively wrote the manuscript. All authors reviewed the manuscript and approved the final version for submission.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Nagao, Y., Mimura, N., Takeda, J. et al. Genetic and transcriptional landscape of plasma cells in POEMS syndrome. Leukemia 33, 1723–1735 (2019). https://doi.org/10.1038/s41375-018-0348-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0348-x
This article is cited by
-
POEMS syndrome remains a mystery after 40 years
Leukemia (2021)
-
A highly heterogeneous mutational pattern in POEMS syndrome
Leukemia (2021)
-
Reply to “POEMS syndrome remains a mystery after 40 years” (Wang Chen. 20-LEU-1856)
Leukemia (2021)