Abstract
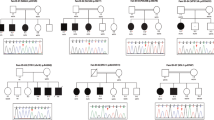
F-box protein 11 (FBXO11) is a member of F-Box protein family, which has recently been proved to be associated with intellectual developmental disorder with dysmorphic facies and behavioral abnormalities (IDDFBA, OMIM: 618089). In this study, 12 intellectual disability individuals from 5 Chinese ID families were collected, and whole exome sequencing (WES), sanger sequencing, and RNA sequencing (RNA-seq) were conducted. Almost all the affected individuals presented with mild to severe intellectual disability (12/12), global developmental delay (10/12), speech and language development delay (8/12) associated with a range of alternate features including increased body weight (7/12), short stature (6/12), seizures (3/12), reduced visual acuity (4/12), hypotonia (1/12), and auditory hallucinations and hallucinations (1/12). Distinguishingly, malformation was not observed in all the affected individuals. WES analysis showed 5 novel FBXO11 variants, which include an inframe deletion variant, a missense variant, two frameshift variants, and a partial deletion of FBXO11 (exon 22–23). RNA-seq indicated that exon 22–23 deletion of FBXO11 results in a new mRNA structure. Conservation and protein structure prediction demonstrated deleterious effect of these variants. The DEGs analysis revealed 148 differentially expressed genes shared among 6 affected individuals, which were mainly associated with genes of muscle and immune system. Our research is the first report of FBXO11-associated IDDFBA in Chinese individuals, which expands the genetic and clinical spectrum of this newly identified NDD/ID syndrome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author (BT), on reasonable request.
References
Totsika V, Liew A, Absoud M, Adnams C, Emerson E. Mental health problems in children with intellectual disability. Lancet Child Adolesc health. 2022;6:432–44.
Hatton C, Glover G, Emerson E, Brown I. People with learning disabilities in England. London: Public Health England. 2018. Available at https://www.gov.uk/government/publications/people-with-learning-disabilities-in-england.
Anderson LL, Larson SA, MapelLentz S, Hall-Lande J. A Systematic Review of U.S. Studies on the Prevalence of Intellectual or Developmental Disabilities Since 2000. Intellect Dev Disabilities. 2019;57:421–38.
Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retardation Dev Disabilities Res Rev. 2002;8:117–34.
Gupta N. Deciphering Intellectual Disability. Indian J Pediatrics. 2023;90:160–67.
Curry CJ, Stevenson RE, Aughton D, Byrne J, Carey JC, Cassidy S, et al. Evaluation of mental retardation: recommendations of a Consensus Conference: American College of Medical Genetics. Am J Med Genet. 1997;72:468–77.
Marti M, Millan MIP, Young JI, Walz K. Intellectual disability, the long way from genes to biological mechanisms. J Transl Genet Genomics. 2020;4:104–13.
Kochinke K, Zweier C, Nijhof B, Fenckova M, Cizek P, Honti F, et al. Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules. Am J Hum Genet. 2016;98:149–64.
Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–18.
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–47.
Study DDD. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–38.
Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–07.
Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607.
Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873.
Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32.
Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–54.
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–56.
Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–80.
Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002.
Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–51.
Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93.
Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73.
Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–804.
Wen X, Li S, Guo M, Liao H, Chen Y, Kuang X, et al. miR-181a-5p inhibits the proliferation and invasion of drug-resistant glioblastoma cells by targeting F-box protein 11 expression. Oncol Lett. 2020;20:235.
Shao L, Zhang X, Yao Q. The F-box protein FBXO11 restrains hepatocellular carcinoma stemness via promotion of ubiquitin-mediated degradation of Snail. FEBS Open Bio. 2020;10:1810–20.
Schieber M, Marinaccio C, Bolanos LC, Haffey WD, Greis KD, Starczynowski DT, et al. FBXO11 is a candidate tumor suppressor in the leukemic transformation of myelodysplastic syndrome. Blood Cancer J. 2020;10:98.
Fritzen D, Kuechler A, Grimmel M, Becker J, Peters S, Sturm M, et al. De novo FBXO11 mutations are associated with intellectual disability and behavioural anomalies. Hum Genet. 2018;137:401–11.
Gregor A, Sadleir LG, Asadollahi R, Azzarello-Burri S, Battaglia A, Ousager LB, et al. De Novo Variants in the F-Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. Am J Hum Genet. 2018;103:305–16.
Jansen S, van der Werf IM, Innes AM, Afenjar A, Agrawal PB, Anderson IJ, et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur J Hum Genet: EJHG. 2019;27:738–46.
Gregor A, Meerbrei T, Gerstner T, Toutain A, Lynch SA, Stals K, et al. De novo missense variants in FBXO11 alter its protein expression and subcellular localization. Hum Mol Genet. 2022;31:440–54.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
San Lucas FA, Wang G, Scheet P, Peng B. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012;28:421–22.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Tan B, Liu S, Feng X, Pan X, Qian G, Liu L, et al. Expanding the mutational and clinical spectrum of Chinese intellectual disability patients with two novel CTCF variants. Front Pediatrics. 2023;11:1195862.
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30.
Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Griffin AM, Cleveland HH, Schlomer GL, Vandenbergh DJ, Feinberg ME. Differential Susceptibility: The Genetic Moderation of Peer Pressure on Alcohol Use. J Youth Adolescence. 2015;44:1841–53.
Lee CG, Seol CA, Ki CS. The first familial case of inherited intellectual developmental disorder with dysmorphic facies and behavioral abnormalities (IDDFBA) with a novel FBXO11 variant. Am J Med Genet Part A. 2020;182:2788–92.
Silva RG, Dupont J, Silva E, Sousa AB. New ocular findings in a patient with a novel pathogenic variant in the FBXO11 gene. J AAPOS. 2022;26:268–70.
Muñoz-Escobar J, Kozlov G, Gehring K. Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding. Protein Sci. 2017;26:2092–97.
Acknowledgements
The author would like to thank all the affected individuals and their families for participating in the study.
Funding
This work was supported by the Scientific and Technological Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202200420) and Program for Youth Innovation in Future Medicine, Chongqing Medical University (W0122).
Author information
Authors and Affiliations
Contributions
Conceptualization: XP and BT; Data curation: XP, LL, XZ and GQ; Formal Analysis: HQ and SHL; Funding acquisition: HY, XD and BT; Investigation: HQ and LL; Project administration: HY, XD and BT, Supervision: HY, XD and BT; Visualization: HQ and SHL; Writing – original draft: HQ; Writing – review & editing: HY, XD, XT and BT.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study is approved by Ethics Committee of The Second Affiliated Hospital of Chongqing Medical University. Written informed consent was provided by the participants’ legal guardians/next of kin.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, X., Liu, L., Zhang, X. et al. FBXO11 variants are associated with intellectual disability and variable clinical manifestation in Chinese affected individuals. J Hum Genet (2024). https://doi.org/10.1038/s10038-024-01255-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s10038-024-01255-4