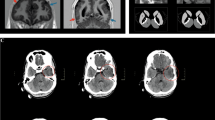
Abstract
The human SET nuclear proto-oncogene (SET) gene is a protein-coding gene that encodes proteins that affects chromatin remodeling and gene transcription. Mutations in the SET gene have been reported to cause intellectual disability (ID) and epilepsy. In this study, we collected and analyzed clinical, genetic, and transcript features of two unrelated Chinese patients with ID. Both patients were characterized by moderate intellectual disability. Whole-exome sequencing identified two novel heterozygous mutations in the SET gene: NM_001122821.1:c.532-3 T > A and NM_001122821.1:c.3 G > C (p.0?). Additionally, RNA sequencing revealed widespread dysregulation of genes involved in NF-kB signaling and neuronal system in these two patients. To our knowledge, this is the first report of SET mutations causing ID in the Chinese population, broadening the genetic and ethnic spectrum of SET-related disorders and highlighting the importance of screening for SET gene variants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author, (BT), on reasonable request.
References
Moeschler JB, Shevell M, Committee on G. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134:e903–18.
Patel DR, Cabral MD, Ho A, Merrick J. A clinical primer on intellectual disability. Transl Pediatr. 2020;9:S23–35.
Baker K, Devine RT, Ng-Cordell E, Raymond FL, consortium I-I, Hughes C. Childhood intellectual disability and parents’ mental health: integrating social, psychological and genetic influences. Br J Psychiatry. 2021;218:315–22.
Jarvela I, Maatta T, Acharya A, Leppala J, Jhangiani SN, Arvio M, et al. Exome sequencing reveals predominantly de novo variants in disorders with intellectual disability (ID) in the founder population of Finland. Hum Genet. 2021;140:1011–29.
Pavinato L, Trajkova S, Grosso E, Giorgio E, Bruselles A, Radio FC, et al. Expanding the clinical phenotype of the ultra-rare Skraban-Deardorff syndrome: two novel individuals with WDR26 loss-of-function variants and a literature review. Am J Med Genet A. 2021;185:1712–20.
Brunet T, Jech R, Brugger M, Kovacs R, Alhaddad B, Leszinski G, et al. De novo variants in neurodevelopmental disorders-experiences from a tertiary care center. Clin Genet. 2021;100:14–28.
Vetrini F, McKee S, Rosenfeld JA, Suri M, Lewis AM, Nugent KM, et al. De novo and inherited TCF20 pathogenic variants are associated with intellectual disability, dysmorphic features, hypotonia, and neurological impairments with similarities to Smith-Magenis syndrome. Genome Med. 2019;11:12.
Stevens SJC, van der Schoot V, Leduc MS, Rinne T, Lalani SR, Weiss MM, et al. De novo mutations in the SET nuclear proto-oncogene, encoding a component of the inhibitor of histone acetyltransferases (INHAT) complex in patients with nonsyndromic intellectual disability. Hum Mutat. 2018;39:1014–23.
Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271:11059–62.
von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–55.
Liu L, Feng X, Liu S, Zhou Y, Dong X, Yao H, et al. Whole-genome sequencing combined RNA-sequencing analysis of patients with mutations in SET binding protein 1. Front Neurosci. 2022;16:980000.
Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772.
Zhang L, Gao J, Liu H, Tian Y, Zhang X, Lei W, et al. Pathogenic variants identified by whole-exome sequencing in 43 patients with epilepsy. Hum Genom. 2020;14:44.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Liu X, Li C, Mou C, Dong Y, Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122.
Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81.
Yuan X, Bai J, Zhang J, Yang L, Duan J, Li Y, et al. CONDEL: detecting copy number variation and genotyping deletion zygosity from single tumor samples using sequence data. IEEE/ACM Trans Comput Biol Bioinform. 2020;17:1141–53.
Steinhaus R, Proft S, Schuelke M, Cooper DN, Schwarz JM, Seelow D. MutationTaster2021. Nucleic Acids Res. 2021;49:W446–51.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37:235–41.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–48.e24.
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6:e1001025.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Griffin AM, Cleveland HH, Schlomer GL, Vandenbergh DJ, Feinberg ME. Differential susceptibility: the genetic moderation of peer pressure on alcohol use. J Youth Adolesc. 2015;44:1841–53.
Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Riggs ER, Bingaman TI, Barry CA, Behlmann A, Bluske K, Bostwick B, et al. Clinical validity assessment of genes frequently tested on intellectual disability/autism sequencing panels. Genet Med. 2022;24:1899–908.
Marinakis NM, Svingou M, Veltra D, Kekou K, Sofocleous C, Tilemis FN, et al. Phenotype-driven variant filtration strategy in exome sequencing toward a high diagnostic yield and identification of 85 novel variants in 400 patients with rare Mendelian disorders. Am J Med Genet A. 2021;185:2561–71.
Richardson R, Splitt M, Newbury-Ecob R, Hulbert A, Kennedy J, Weber A, et al. SET de novo frameshift variants associated with developmental delay and intellectual disabilities. Eur J Hum Genet. 2018;26:1306–11.
Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Zhang X, Wu Y, Duan X, Chen W, Zou H, Zhang M, et al. Upregulation of SET expression by BACE1 and its implications in Down syndrome. Mol Neurobiol. 2015;51:781–90.
Li J, He Q, Wang L, Chen D, Qiu C, Xu P, et al. SET knockdown attenuated phenotype modulation and calcium channel associated markers of airway smooth muscle cells in asthmatic mice. Ann Transl Med. 2021;9:657.
Acknowledgements
We appreciate the patients and their family members for their participation in this study.
Funding
This work was supported by the Scientific and Technological Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202200420), Nan’an District Science and Health Joint Medical Scientific Research Project (2020-01), Program for Youth Innovation in Future Medicine, Chongqing Medical University (W0122), and Maternal and Child Health Research Cultivation Project of the Chongqing Health Commission (2023FY201).
Author information
Authors and Affiliations
Contributions
Conceptualization: XP, SL, XF and BT; Data curation: XP; Formal Analysis: SL, XF and BT; Funding acquisition: HY, XD and BT; Investigation: LL, ZX, GQ and NL; Project administration: HY, XD and BT, Supervision: HY, XD and BT; Visualization: SL, XF and BT; Writing – original draft: SL, XF and BT; Writing – review & editing: HY, XD and BT.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, X., Liu, S., Feng, X. et al. Whole exome sequencing and transcriptome analysis in two unrelated patients with novel SET mutations. J Hum Genet 68, 867–874 (2023). https://doi.org/10.1038/s10038-023-01196-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01196-4