Abstract
T-cell immunodeficiency may pose a diagnostic challenge to clinicians, especially when the basic T-cell immune workup is not sufficiently informative. An intensive assessment of thymus capabilities that involves either measuring the recent thymic emigrant cells or analyzing the T-cell receptor (TCR) repertoire is often required to estimate the severity and nature of the immune disorder. A comprehensive T-cell immune workup, including TCR excision circles (TRECs) and TCR repertoire analyses, was performed in three patients with various degrees of severity of T-cell immunodeficiency. All three patients had normal peripheral CD3+ T lymphocytes. TCR repertoire analysis revealed oligoclonal (patient 1), restricted (patient 2), and near-normal (patient 3) patterns. TREC quantification was significantly reduced in patients 1 and 2 but normal in patient 3. Based on clinical features at presentation and at follow-up, and supported by the results of immunologic studies, patients 1 and 2 were diagnosed as having significant T-cell immunodeficiency and patient 3 as having T-cell immunocompetence. Assessment of thymus capabilities by TRECs and TCR repertoire analyses is helpful in diagnosing patients with T-cell immunodeficiency and should be part of the evaluation of every patient suspected of having that condition.
Similar content being viewed by others
Main
Primary immunodeficiencies represent a heterogeneous group of inherited diseases characterized by a defect in one of the components that compose the immune system. T-cell immunodeficiency is probably the most severe form, with most affected individuals being especially vulnerable to serious viral, fungal, and opportunistic infections within the first few months of life (1). The spectrum of T-cell immunodeficiency varies from severe combined immunodeficiency (SCID), which requires restoration of the immune system by allogeneic hematopoietic stem cell transplantation, to milder types that are characterized by having a favorable outcome, even without treatment (2). Patients are often diagnosed by clinical features at presentation and family history, supplemented by immunologic evaluation. Reduced numbers of lymphocyte subsets, including T, B, and natural killer cells in the peripheral blood and a depressed response of T cells to mitogen or antigen stimulation are typically seen in the severe types of the condition. These tests are not informative enough to lead to the correct diagnosis in some patients, particularly those with a milder phenotype. Therefore, an intensive workup of the thymus, which is the main organ for T-cell development and maturation, is often required (3). T-cell maturation in the thymus goes through distinct stages defined phenotypically by the expression of CD4 and CD8 coreceptors and rearrangement of DNA segments [Variable, Diversity, and Joining (V-D-J)] to form a functional T-cell receptor (TCR). Accurate equipment involving recombination, activation, DNA damage recognition, and subsequent gene repair is critical for the development of the different TCRs (4,5). Analyzing the products of this process, including the expression of the TCR repertoire and the quantification of excised DNA circles [TCR excision circles (TRECs)] is indicative of robust T-cell immunity (6,7). Indeed, performing analyses to demonstrate the expression of the TCR repertoire is highly important, particularly when exaggerated expansion of a few T-cell clones results in misleading numbers of T cells (8). The presence of TRECs, which are not replicated during peripheral T-cell division, serves to identify new thymus-derived T cells (9). In this study, we show the utility of these tests in the evaluation of three patients with different degrees of severity of T-cell immunodeficiency. Importantly, the findings of these tests were closely correlated with disease severity and outcome.
METHODS
Patients.
Patients suspected of having T-cell immunodeficiency based on clinical findings, family history, and immune evaluations according to criteria of the World Health Organization (10) were eligible for this study. HIV was excluded by PCR. All entrants gave their signed informed consent to participate in this study that was approved by the Institutional Review Board (Sheba Medical Center, Tel Hashomer).
Immune function.
Cell surface markers of peripheral blood mononuclear cells (PBMCs), using immunofluorescent staining and flow cytometry (Epics V; Coulter Electronics, Hialeah, FL) with antibodies purchased from Coulter Diagnostics, and lymphocyte proliferation in response to phytohemagglutinin and anti-CD3, using tritiated thymidine incorporation were determined as previously described (11). Age-adjusted normal values for the cell surface markers are given in Table 2. For lymphocyte proliferation studies, the cells were harvested 3 d after collection, and samples were counted in a liquid scintillation counter. All assays were performed in triplicates, and a stimulation index was calculated as the ratio between stimulated and unstimulated lymphocyte responses. The resultant stimulation index was compared with the stimulation index obtained from normal controls. Serum concentration of immunoglobulins was measured by nephelometry.
Quantitation of TCR genes.
Representatives of specific TCR Vβ families were detected and quantified using flow cytometry (Becton-Dickinson, Calibur) and referred to the Quick Reference Card according to manufacturer's (Beckman Coulter) instructions. Based on our long-term experience with this assay, we defined an oligoclonal pattern when at least one Vβ-chain was responsible for >20% of all T cells in the absence of other Vβ chains, a restricted pattern when fewer than 10 Vβ chains were expressed, and a normal pattern when most of the Vβ chains were normally expressed.
TCR-γ rearrangements were amplified by PCR according to the standardized Biomed 2 protocol using four primers designed to cover the possible sequences of different variable region genes and their consensus sequences (Vγ 9/2, Vγ 11, Vγ f1, and Vγ 10/2). The reverse primer for these reactions is common to all four forward primers and is a consensus primer for the joining region (12). For GeneScan analyses, the four Vγ primers were fluorescently labeled (FAM or HEX fluorochrome) and used in PCRs as described in the Biomed 2 protocol (12). Fluorescence-labeled PCR amplificate (1 μL of each) was added to a mixture of 8.5 μL deionized formamide and 0.5 μL GeneScan 500TM Rox internal standard (PE Applied Biosystem, Weiterstadt, Germany). Figure 2 (panel A) displays representative examples of TCR-γ spectratyping to illustrate each of the three possible assay interpretations, i.e. oligoclonal, restricted, and normal (Gaussian) distribution.
TCR-γ spectratyping. A, representative examples of TCR-γ spectratyping (Genescan) of fluorescence-labeled PCR to illustrate each of the three possible assay interpretations: oligoclonal (left panel), restricted distribution (middle panel), and normal (Gaussian) distribution (right panel). B–E, TCR-γ spectratyping using four consensus Vγ primers (Vγ 9/2, Vγ 11, Vγ f1, and Vγ 10/2) for the characterization of the TCR-γ in the three patients' PBMC (B–D) and healthy control (E). Panel B represents patient 1's clonal profile with a dominant Vγ 9/2 clone, panel C, represents patient 2's oligoclonal profile without clearly dominant clones, and panel D represents patient 3's profile with expansion of a Vγ 11 clone.
Analysis of TRECs.
This analysis was performed by using DNA extracted from the study patients' PBMCs. The amount of signal joint (sj) TREC copies per DNA content was determined by real-time quantitative PCR as previously described (11,13). In brief, genomic DNA was isolated from PBMCs after Ficoll-Hypaque gradient centrifugation and extracted with a Promega Wizard genomic purification kit (Promega, Madison, WI) according to the manufacturer's instructions. The following primers and probe were used for TREC PCR: sj-5′ forward: cacatccctttcaaccatgct (900 nM); sj-3′ reverse: gccagctgcagggtttagg (900 nM), and the oligo 5′ FAM-acacctctggtttttgtaaaggtgcccact-TAMRA p-3′ (250 nM) as a detection probe. Amplification reactions (25 μL) contained 0.5 μg of genomic DNA, 12.5 μL of TaqMan universal PCR master mix (Perkin Elmer Applied Biosystem, Foster City, CA, USA), and the appropriate primers and probes. PCR (2 min at 50°C followed by 95°C for 10 min, then 40 cycles at 95°C for 15 s and 60°C for 1 min) was carried out in the ABI PRISM 7900 Sequence Detector TaqMan system (Applied Biosystems, Rotkreuz, Switzerland). The number of TRECs in a given sample was estimated by comparing the cycle threshold value obtained with a standard curve obtained from PCRs performed with 10-fold serial dilutions of an internal standard kindly provided by Dr. Daniel Douek (Vaccine Research Center, National Institute of Allergy and Infectious Diseases, Bethesda, MD). The dilutions contained between 106 and 103 copies of sjTRECs, and three reactions were run with each dilution. Amplification of β-actin was used to verify the presence of genomic DNA. PCR for β-actin used identical cycles with 1 μL of DNA, 375 nmol/L primers (F, 5′-tcacccacactgtgcccatctacgag; R, 5′-cagcgaaccgctcattgccatgg), and 250 nmol/L probe (FAM-5′-atgccctcccccatgccatcctgcgt-TAMRA). Fifty age-matched normal individuals for whom the diagnosis of primary immunodeficiency was excluded were used as controls.
RESULTS
Clinical features.
There was a history of parental consanguinity for all three patients. Patient 1's growth and development was normal and she exhibited no symptoms suggestive of immunodeficiency until the age of 7 mo, when she developed progressive respiratory symptoms. Failure to thrive was noticed either at the time of diagnosis or shortly thereafter. Physical examination revealed reduced-sized tonsils and cervical lymph nodes (Table 1). She was diagnosed with interstitial Pneumocystis carinii pneumonia and cytomegalovirus (CMV) and later developed chronic Rota virus infection. She received mismatched related bone marrow transplantation when she was 10 mo of age, and currently, she is alive and well.
Patient 2 started to suffer from recurrent chest infections complicated with bronchiectasis development around 1 y of age. She later had oral thrush, recurrent skin infections, and life-threatening viral infections. Physical examination revealed reduced-sized tonsils and cervical lymph nodes. She underwent related identical donor bone marrow transplantation when she was 4.3 y of age, and currently, she is alive and well.
Patient 3 presented with failure to thrive and respiratory symptoms at the age of 6 mo. Physical examination revealed normal-sized tonsils and cervical lymph nodes. CMV was isolated from her blood by PCR detection, and IgG serology was positive later on. She underwent a lung biopsy that failed to detect inclusion bodies. She recovered from CMV infection without receiving any specific treatment and currently, at 15 mo of age, is in apparent good health, gaining weight, and shows no further symptoms suggestive of immunodeficiency.
Immunologic studies.
All three patients had normal peripheral CD3+ T lymphocytes (Table 2). Subsets of CD4+ T lymphocytes and CD19+ B lymphocytes were slightly reduced in patients 1 and 2. These patients also had evidence of humoral immunodeficiency as evidenced by the lack of immunoglobulins. In vitro T lymphocyte responses to phytohemagglutinin was significantly reduced in patient 1, moderately reduced in patient 2, and slightly reduced in patient 3. Patients 1 and 2 also had reduced responses to CD3 antibody, whereas patient 3 had a normal response.
Thymus capabilities.
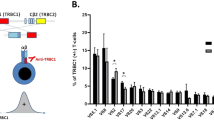
The TCR-Vβ region in the CD3+ cells the three patients was examined by FACS (Fig. 1). Patient 1 had an oligoclonal profile with dominant Vβ 11, 13.1, 13.2, 13.6 clones and lack of the 17 other Vβs that were tested. Patient 2 had restriction representation with 11 different reduced TCR-Vβs (1, 5.1, 5.2, 5.3, 7.2, 9, 12, 16, 20, 21.3, 22) but no clear dominant clones. Patient 3 had a polyclonal TCR profile with representation of all Vβs tested, but there was expansion of the Vβ3 receptor. To better define the lymphoid clonality of these patients, we also performed PCR analysis of the TCR-γ gene rearrangement (Fig. 2), which represents the “prototype” of restricted repertoire targets (14). Similar to the pattern obtained from analysis of their TCR-Vβ, patient 1 had a clonal repertoire of the TCR-γ gene (Fig. 2panel B), whereas patient 2 had a more restricted repertoire without dominant clones (Fig. 2panel C). Patient 3 had a few dominant clones with slight restriction of other TCR-γ gene rearrangements (Fig. 2panel D). For example, although healthy controls TCR-γ 9/2 gives a normal distribution (Fig. 2panel E), patients 1 and 2 showed only one peak, and patient 3 has several peaks in their TCR-γ 9/2 gene rearrangements.
TCR-Vβ repertoire. Relative expression of the various Vβ families in our three patients' CD3+ cells (black bar) compared with normal control data obtained from the IOTest beta Mark PN IM3497—Quick Reference Card. The minimum, maximum, and SD values are given for each Vβ. Panel A represents patient 1's oligoclonal profile with a dominant Vβ 11, 13.1, 13.2, 13.6 clones, panel B represents patient 2's restriction representation with absence of 12 different TCR-Vβ but with no clearly dominant clones, and panel C represents patient 3's profile with expansion of a Vβ 3.
The amount of recent thymic emigrant cells as determined by real-time PCR analysis of TREC copies per 0.5 μg of DNA obtained from the three patients' PBMCs was significantly reduced in patients 1 (undetectable copies) and 2 (50 copies), compared with 50 healthy age-matched controls. The undetectable level of TRECs seen in patient 1 was similar to what has been reported in patients with SCID (13). In contrast, patient 3 had 480 copies of TRECs per 0.5 μg of DNA, a result similar to that of normal age-matched controls (Table 2).
DISCUSSION
T-cell immunodeficiencies have a wide spectrum. Because treatment differs considerably according to the severity of the disease, establishing the correct diagnosis supported by a thorough evaluation of the T-cell immunity is vitally important (15). The diagnosis of T-cell immunodeficiency is usually straightforward and should be confirmed by molecular genetic analysis when a patient is clinically immunodeficient and lab tests reveal an absence of T lymphocytes (16). Significant T-cell immunodeficiency should, however, be considered in clinically immunodeficient patients who present with an atypical immunologic phenotype as a result of residual autologous T cells (17,18), maternal T-cell engraftment (19), a leaky thymus with release of some T-cell clones (11), or of being part of combined immunodeficiency syndrome. In cases such as these, elaborate and intensive studies of the thymus, including assessment of recent thymic emigrant cells or analysis of the TCR repertoire, are required to estimate the severity and nature of the immune disorder (20). Indeed, in the three cases presented herein, these assays were able to distinguish between patients with normal and abnormal T-cell immunity regardless of the normal numbers of lymphocytes/T cells that were found in each of them. Patients 1 and 2 had clear-cut evidence of humoral immunodeficiency, including the lack of immunoglobulins (both patients), reduced B lymphocytes (patient 1), and bronchiectasis (patient 2), and were therefore diagnosed as having combined immunodeficiency. In patient 1, maternal T-cell engraftment and Omenn syndrome features were excluded, and so we assumed that her T cells originated from residual autologous ones with the release of some T-cell clones. The older age at presentation of patient 2 is probably enough to exclude the possibility of SCID, however, the findings of her clinical history and immune workup, including abnormal thymus capabilities, are indicative of combined immunodeficiency. A restricted pattern of TCR repertoire and reduced TRECs has, to our knowledge, never been described in such patients, and it emphasizes the utility of these tests in diagnosing “older” patients suspected of having combined immunodeficiency. In contrast, patient 3, who had normal TRECs and a nonclonal TCR repertoire, was diagnosed as having normal T-cell immunity, despite initial clinical evidence of T-cell immunodeficiency and slightly reduced T-cell response to phytohemagglutinin mitogenic stimulation. In addition, she had normal immunoglobulin levels and positive specific antibody response to antigen (CMV). Indeed, her outcome turned out to be favorable even without specific treatment. Because the follow-up of this patient is short, further studies to determine thymus capabilities will be required to exclude the later development of significant T-cell immunodeficiency.
The level of T-cell rearrangement excision circles in peripheral blood T cells is measured to quantify recent thymic emigrant cells (6). These are nonreplicating episomal DNA byproducts of the final step of TCR rearrangement that are present in newly emigrated thymic CD4 and CD8 T cells. Because TRECs are exclusively derived from T cells, patients with dysplastic thymus and profound T-cell immunodeficiency will obviously have low copies of TRECs. However, we now demonstrate that this test is also valuable in patients with residual T cell numbers (patients 1 and 2) and even in those with some T-cell function (patient 2). In the past, TRECs were suggested as a stable analyte that can identify T-cell lymphopenia in dried blood spots obtained from newborn screening (21). It is considered to be an accurate way to assess immune reconstitution after hematopoietic stem cell transplantation and to correlate with outcome (12,22). In addition, it may detect thymic dysfunction in patients with CVID (23) or be used to evaluate HIV patients and their response to antiretroviral therapy (24). Assessing the numbers of T cells expressing CD4+CD45RA+ is another tool to follow naïve T cells, however this measurement is not as accurate as TRECs quantification, because CD4+CD45RA+ may remain quiescent for an extended period in the periphery, they may proliferate in an antigen-independent manner, and they may rapidly convert to CD45RO+ memory/effector T cells.
Analysis of the TCR is valuable in the diagnosis of certain T-cell deficiencies, particularly those in which there is expansion of certain T-cell clones evoked by malignancy or repeated exposure to a specific antigen or super antigen. The result of this process is expansion of T cells of one or more variable segments and a skewed repertoire, such as that seen in patients with Omenn syndrome (25). We used two different modalities to assess the TCR repertoire in our patients. Both modalities are usually performed in specialized research laboratories, and the results are interpreted by a molecular immunologist. Although the TCR-Vβ assay also includes referred normal control values in the manufacturer's kit, the TCR-Vγ assay usually requires an experienced interpreter. The TCR-Vβ assay measures the relative expression of the functional variable segment of the variable β chain on CD3+ cells. This flow cytometry assay is performed more frequently in clinical practice, has well-established controls, and can easily be interpreted. It does, however, require a fresh blood sample, a feature that limits its use. The γ-chain gene spectratyping assay examines the clonality of the TCR-Vγ by fluorochrome-labeled PCR. Although it is typically not expressed on the cell surface, TCR-Vγ remains rearranged in the T-cell genome and provides a conventional marker of clonality due to its presence in both TCRαβ and TCRγδ T cells (26,27). In this assay, four consensus Vγ primers are used to allow for a rapid and reagent-efficient assessment of clonality. The results derived from this assay are not as clear as those of the TCRβ assay, although qualified labs can perform this analysis very accurately and sometimes do so in conjunction with the TCR-Vβ assay. Our results showed that similar interpretations were obtained by analyzing both the expression of the TCR-Vβ and the DNA analysis of the γ-chain gene, indicating their value in diagnosing T-cell immunodeficiency. Importantly, we found that diminished use of certain segments might lead to a skewed TCR distribution, even in patients with immunologic immaturity. In patient 3, we assume that the clonal expansion of the TCR may reflect recent exposure to a pathogen(s) or skewing toward self antigens. The latter is less likely because the patient did not show any evidence of autoimmunity.
Taken together, we believe that assessment of thymus capabilities by TRECs and the TCR repertoire is important in patients with T-cell immunodeficiency and that it should be added to the measurement of lymphocyte subsets and response to mitogenic or antigenic stimulation in the evaluation of every patient undergoing a workup for T-cell immunodeficiency.
Abbreviations
- PBMCs:
-
peripheral blood mononuclear cells
- SCID:
-
severe combined immunodeficiency
- TCR:
-
T-cell receptor
- TRECs:
-
T-cell receptor excision circles
References
Buckley RH 2004 Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol 22: 625–655
Roifman CM 2006 Approach to the diagnosis of severe combined immunodeficiency. In: Barbara D, Robert GH, James D (eds) Manual of Molecular and Clinical Laboratory Immunology. ASM Press, Washington, DC, pp 895–890
Roifman CM 2005 Studies of patients' thymi aid in the discovery and characterization of immunodeficiency in humans. Immunol Rev 203: 143–155
Eason DD, Cannon JP, Haire RN, Rast JP, Ostrov DA, Litman GW 2004 Mechanisms of antigen receptor evolution. Semin Immunol 16: 215–226
de Villartay JP, Poinsignon C, de Chasseval R, Buck D, Le Guyader G, Villey I 2003 Human and animal models of V(D)J recombination deficiency. Curr Opin Immunol 15: 592–598
Ye P, Kirschner DE 2002 Measuring emigration of human thymocytes by T-cell receptor excision circles. Crit Rev Immunol 22: 483–497
Hazenberg MD, Verschuren MC, Hamann D, Miedema F, van Dongen JJ 2001 T cell receptor excision circles as markers for recent thymic emigrants: basic aspects, technical approach, and guidelines for interpretation. J Mol Med 79: 631–640
Zhang J, Quintal L, Atkinson A, Williams B, Grunebaum E, Roifman CM 2005 Novel RAG1 mutation in a case of severe combined immunodeficiency. Pediatrics 116: e445–e449
Haynes BF, Markert ML, Sempowski GD, Patel DD, Hale LP 2000 The role of the thymus in immune reconstitution in aging, bone marrow transplantation, and HIV-1 infection. Annu Rev Immunol 18: 529–560
Geha RS, Notarangelo LD, Casanova JL, Chapel H, Conley ME, Fischer A, Hammarström L, Nonoyama S, Ochs HD, Puck JM, Roifman C, Seger R, Wedgwood J 2007 International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee. J Allergy Clin Immunol 120: 776–794
Roifman CM, Somech R, Grunebaum E 2008 Matched unrelated bone marrow transplant for T+ combined immunodeficiency. Bone Marrow Transplant 41: 947–952
van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, García-Sanz R, van Krieken JH, Droese J, González D, Bastard C, White HE, Spaargaren M, González M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA 2003 Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17: 2257–2317
Douek DC, Vescio RA, Betts MR, Brenchley JM, Hill BJ, Zhang L, Berenson JR, Collins RH, Koup RA 2000 Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet 355: 1875–1881
Theodorou I, Raphaël M, Bigorgne C, Fourcade C, Lahet C, Cochet G, Lefranc MP, Gaulard P, Farcet JP 1994 Recombination pattern of the TCR gamma locus in human peripheral T-cell lymphomas. J Pathol 174: 233–242
Fischer A 2002 Primary immunodeficiency diseases: natural mutant models for the study of the immune system. Scand J Immunol 55: 238–241
Somech R, Roifman CM 2005 Mutation analysis should be performed to rule out gammac deficiency in children with functional severe combined immune deficiency despite apparently normal immunologic tests. J Pediatr 147: 555–557
Schmalstieg FC, Leonard WJ, Noguchi M, Berg M, Rudloff HE, Denney RM, Dave SK, Brooks EG, Goldman AS 1995 Missense mutation in exon 7 of the common gamma chain gene causes a moderate form of X-linked combined immunodeficiency. J Clin Invest 95: 1169–1173
Stephan V, Wahn V, Le Deist F, Dirksen U, Broker B, Muller-Fleckenstein I, Horneff G, Schroten H, Fischer A, de Saint Basile G 1996 Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N Engl J Med 335: 1563–1567
Kumaki S, Ishii N, Minegishi M, Ohashi Y, Hakozaki I, Nonoyama S, Imai K, Morio T, Tsuge I, Sakiyama Y, Miyanoshita A, Miura J, Mayumi M, Heike T, Katamura K, Takada H, Izumi I, Kamizono J, Hibi S, Sasaki H, Kimura M, Kikuta A, Date Y, Sako M, Tanaka H, Sano K, Sugamura K, Tsuchiya S 2000 Characterization of the gammac chain among 27 unrelated Japanese patients with X-linked severe combined immunodeficiency (X-SCID). Hum Genet 107: 406–408
Somech R, Somers GR, Chitayat D, Grunebaum E, Atkinson A, Kolomietz E, Roifman CM 2008 Fatal lung fibrosis associated with immunodeficiency and gonadal dysgenesis in 46XX sisters—a new syndrome. Am J Med Genet A 146A: 8–14
Chan K, Puck JM 2005 Development of population-based newborn screening for severe combined immunodeficiency. J Allergy Clin Immunol 115: 391–398
Guggenheim R, Somech R, Grunebaum E, Atkinson A, Roifman CM 2006 Bone marrow transplantation for cartilage-hair-hypoplasia. Bone Marrow Transplant 38: 751–756
Guazzi V, Aiuti F, Mezzaroma I, Mazzetta F, Andolfi G, Mortellaro A, Pierdominici M, Fantini R, Marziali M, Aiuti A 2002 Assessment of thymic output in common variable immunodeficiency patients by evaluation of T cell receptor excision circles. Clin Exp Immunol 129: 346–353
De Rossi A, Walker AS, De Forni D, Klein N, Gibb DM 2005 Paediatric European Network for Treatment of AIDS. Relationship between changes in thymic emigrants and cell-associated HIV-1 DNA in HIV-1-infected children initiating antiretroviral therapy. Antivir Ther 10: 63–71
Pirovano S, Mazzolari E, Pasic S, Albertini A, Notarangelo LD, Imberti L 2003 Impaired thymic output and restricted T-cell repertoire in two infants with immunodeficiency and early-onset generalized dermatitis. Immunol Lett 86: 93–97
van Krieken JH, Langerak AW, Macintyre EA, Kneba M, Hodges E, Sanz RG, Morgan GJ, Parreira A, Molina TJ, Cabeçadas J, Gaulard P, Jasani B, Garcia JF, Ott M, Hannsmann ML, Berger F, Hummel M, Davi F, Brüggemann M, Lavender FL, Schuuring E, Evans PA, White H, Salles G, Groenen PJ, Gameiro P, Pott Ch, Dongen JJ 2007 Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 21: 201–206
Nikolich-Zugich J, Slifka MK, Messaoudi I 2004 The many important facets of T-cell repertoire diversity. Nat Rev Immunol 4: 123–132
Acknowledgements
We thank Esther Eshkol for editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amariglio, N., Lev, A., Simon, A. et al. Molecular Assessment of Thymus Capabilities in the Evaluation of T-Cell Immunodeficiency. Pediatr Res 67, 211–216 (2010). https://doi.org/10.1203/PDR.0b013e3181c6e554
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e3181c6e554
This article is cited by
-
SLP76 Mutation Associated with Combined Immunodeficiency and EBV-Related Lymphoma
Journal of Clinical Immunology (2023)
-
Inadequate Activation of γδT- and B-cells in Patient with Wiskott-Aldrich Syndrome (WAS) Portrayed by TRG and IGH Repertoire Analyses
Journal of Clinical Immunology (2023)
-
Immune and TRG repertoire signature of the thymus in Down syndrome patients
Pediatric Research (2021)
-
MHC II deficient infant identified by newborn screening program for SCID
Immunologic Research (2018)
-
Co-appearance of OPV and BCG vaccine-derived complications in two infants with severe combined immunodeficiency
Immunologic Research (2018)