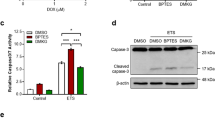
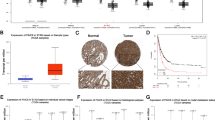
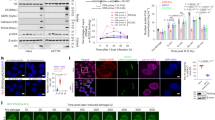
Abstract
SALL4 has recently been identified to promote chemo-resistance in multiple types of cancer, but the underlying mechanism remains to be fully established. Open chromatin structure is important for DNA damage response (DDR) and DNA repair. Here, we demonstrate that SALL4 promotes open chromatin by destabilizing heterochromatin protein 1α (HP1α) by recruiting ubiquitin E3 ligase CUL4B to HP1α. The silencing of SALL4 in cancer cells decreased the expression levels of Glut1 and inhibited glycolysis in cancer cells. The upregulation of HP1α in human cancer cells suppressed open chromatin, glycolysis and Glut1 expression levels. Therefore, SALL4 promotes the expression of Glut1 and open chromatin through a HP1α-dependent mechanism. Impaired DDR in SALL4-deficient human cancer cells can be rescued by the restored expression of Glut1, indicating the importance of HP1α-Glut1 axis in SALL4-mediated DDR. These findings demonstrate that SALL4 could induce drug resistance by enhancing DDR and DNA repair through promoting glycolysis and subsequent chromatin remodeling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Kim J, Nakasaki M, Todorova D, Lake B, Yuan CY, Jamora C et al. p53 Induces skin aging by depleting Blimp1+ sebaceous gland cells. Cell Death Dis 2014; 5: e1141.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014; 511: 246–250.
Lu X, Mazur SJ, Lin T, Appella E, Xu Y . The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene 2014; 33: 2655–2664.
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 2010; 70: 10433–10444.
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 2006; 8: 1114–1123.
Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 2006; 133: 3005–3013.
Kohlhase J, Heinrich M, Liebers M, Frohlich Archangelo L, Reardon W, Kispert A . Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet Genome Res 2002; 98: 274–277.
Miettinen M, Wang Z, McCue PA, Sarlomo-Rikala M, Rys J, Biernat W et al. SALL4 expression in germ cell and non-germ cell tumors: a systematic immunohistochemical study of 3215 cases. Am J Surg Pathol 2014; 38: 410–420.
Chen YY, Li ZZ, Ye YY, Xu F, Niu RJ, Zhang HC et al. Knockdown of SALL4 inhibits the proliferation and reverses the resistance of MCF-7/ADR cells to doxorubicin hydrochloride. BMC Mol Biol 2016; 17: 6.
Liu L, Zhang J, Yang X, Fang C, Xu H, Xi X . SALL4 as an Epithelial-Mesenchymal Transition and Drug Resistance Inducer through the Regulation of c-Myc in Endometrial Cancer. PLoS One 2015; 10: e0138515.
Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan B et al. SALL4 is a new target in endometrial cancer. Oncogene 2015; 34: 63–72.
Yang M, Xie X, Ding Y . SALL4 is a marker of poor prognosis in serous ovarian carcinoma promoting invasion and metastasis. Oncol Rep 2016; 35: 1796–1806.
Forghanifard MM, Moghbeli M, Raeisossadati R, Tavassoli A, Mallak AJ, Boroumand-Noughabi S et al. Role of SALL4 in the progression and metastasis of colorectal cancer. J Biomed Sci 2013; 20: 6.
Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang M et al. SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene 2014; 33: 5491–5500.
Yanagihara N, Kobayashi D, Kuribayashi K, Tanaka M, Hasegawa T, Watanabe N . Significance of SALL4 as a drugresistant factor in lung cancer. Int J Oncol 2015; 46: 1527–1534.
Yong KJ, Chai L, Tenen DG . Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med 2013; 369: 1171–1172.
Cheng J, Gao J, Shuai X, Tao K . Oncogenic protein SALL4 and ZNF217 as prognostic indicators in solid cancers: a metaanalysis of individual studies. Oncotarget 2016; 7: 24314–24325.
Han SX, Wang JL, Guo XJ, He CC, Ying X, Ma JL et al. Serum SALL4 is a novel prognosis biomarker with tumor recurrence and poor survival of patients in hepatocellular carcinoma. J Immunol Res 2014; 2014: 262385.
Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 2009; 4: 226–235.
Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology 2013; 57: 1469–1483.
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N et al. Drug resistance in cancer: an overview. Cancers 2014; 6: 1769–1792.
Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006; 355: 983–991.
Bouwman P, Jonkers J . The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012; 12: 587–598.
Xiong J, Todorova D, Su NY, Kim J, Lee PJ, Shen Z et al. Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J Cell Biol 2015; 208: 513–520.
Shiloh Y, Ziv Y . The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14: 197–210.
Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol 2009; 11: 92–96.
Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 2006; 8: 870–876.
Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol 2007; 178: 1101–1108.
Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE . Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol 2007; 178: 209–218.
Faucher D, Wellinger RJ . Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet 2010; 6: e1001082.
Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008; 31: 167–177.
Petroski MD, Deshaies RJ . Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6: 9–20.
Sulli G, Di Micco R, d'Adda di Fagagna F . Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 2012; 12: 709–720.
Liu XS, Little JB, Yuan ZM . Glycolytic metabolism influences global chromatin structure. Oncotarget 2015; 6: 4214–4225.
Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 2012; 40: 4794–4803.
Young CD, Lewis AS, Rudolph MC, Ruehle MD, Jackman MR, Yun UJ et al. Modulation of glucose transporter 1 (GLUT1) expression levels alters mouse mammary tumor cell growth in vitro and in vivo. PLoS One 2011; 6: e23205.
Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011; 145: 732–744.
Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010; 140: 280–293.
Lu J, Jeong HW, Kong N, Yang Y, Carroll J, Luo HR et al. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One 2009; 4: e5577.
Molina-Serrano D, Kirmizis A . Beyond the histone tail: acetylation at the nucleosome dyad commands transcription. Nucleus 2013; 4: 343–348.
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C . Clonogenic assay of cells in vitro. Nat Protoc 2006; 1: 2315–2319.
Abmayr SM, Yao T, Parmely T, Workman JL . Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol 2006; Chapter 12: Unit 12 11.
Acknowledgements
We thank Michael Qiu for technical support. This study is supported by the National Natural Science Foundation of China (No. 815300045, 81373166, 81430032), a grant from the National High-tech R&D Program (863 Program No. 2015AA020310), Guangdong Provincial Key Laboratory of Tumor Immunotherapy and Guangzhou Key Laboratory of Tumor Immunology Research, South Wisdom Valley Innovative Research Team Program (2014) No. 365, Major basic research developmental project of the Natural Science Foundation of Guangdong Province, Shenzhen Municipal Science and Technology Innovation Council (20140405201035), and grants from California Institute for Regenerative Medicine (TR3-05559, RT3-07899).
Author contributions
J.K. with the help of S.X. and L.Y. performed experiments. J.K, X.F. and Y.X. planned the experiments and interpreted the data. X.F. and Y.X. provided the administrative support. J.K. and Y.X. were responsible for the initial draft of the manuscript, whereas other authors contributed to the final edited versions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Xu, S., Xiong, L. et al. SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene 36, 6472–6479 (2017). https://doi.org/10.1038/onc.2017.265
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.265
This article is cited by
-
The N6-methyladenosine METTL3 regulates tumorigenesis and glycolysis by mediating m6A methylation of the tumor suppressor LATS1 in breast cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
SALL4 promotes angiogenesis in gastric cancer by regulating VEGF expression and targeting SALL4/VEGF pathway inhibits cancer progression
Cancer Cell International (2023)
-
SALL4 Expression in Epithelial Gastric Cancer: Epigenetic or Epiphenomenon?
Digestive Diseases and Sciences (2023)
-
The miR-1185-2-3p—GOLPH3L pathway promotes glucose metabolism in breast cancer by stabilizing p53-induced SERPINE1
Journal of Experimental & Clinical Cancer Research (2021)
-
Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2
Molecular Cancer (2021)