Abstract
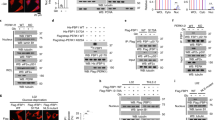
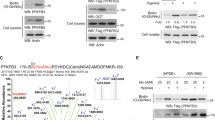
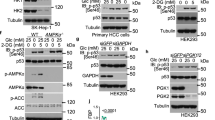
The bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-4 (PFKFB4) controls metabolic flux through allosteric regulation of glycolysis. Here we show that p53 regulates the expression of PFKFB4 and that p53-deficient cancer cells are highly dependent on the function of this enzyme. We found that p53 downregulates PFKFB4 expression by binding to its promoter and mediating transcriptional repression via histone deacetylases. Depletion of PFKFB4 from p53-deficient cancer cells increased levels of the allosteric regulator fructose-2,6-bisphosphate, leading to increased glycolytic activity but decreased routing of metabolites through the oxidative arm of the pentose-phosphate pathway. PFKFB4 was also required to support the synthesis and regeneration of nicotinamide adenine dinucleotide phosphate (NADPH) in p53-deficient cancer cells. Moreover, depletion of PFKFB4-attenuated cellular biosynthetic activity and resulted in the accumulation of reactive oxygen species and cell death in the absence of p53. Finally, silencing of PFKFB4-induced apoptosis in p53-deficient cancer cells in vivo and interfered with tumour growth. These results demonstrate that PFKFB4 is essential to support anabolic metabolism in p53-deficient cancer cells and suggest that inhibition of PFKFB4 could be an effective strategy for cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Lunt SY, Vander Heiden MG . Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011; 27: 441–464.
Kruiswijk F, Labuschagne CF, Vousden KH . p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 2015; 16: 393–405.
Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149: 1269–1283.
Kawauchi K, Araki K, Tobiume K, Tanaka N . p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol 2008; 10: 611–618.
Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653.
Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126: 107–120.
Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 2012; 22: 585–600.
Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol 2011; 13: 310–316.
Jiang P, Du W, Mancuso A, Wellen KE, Yang X . Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013; 493: 689–693.
Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov 2012; 2: 328–343.
Lavin MF, Gueven N . The complexity of p53 stabilization and activation. Cell Death Differ 2006; 13: 941–950.
Ho J, Benchimol S . Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ 2003; 10: 404–408.
Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev 1999; 13: 2490–2501.
Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–1075.
Okar DA, Manzano A, Navarro-Sabate A, Riera L, Bartrons R, Lange AJ . PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem Sci 2001; 26: 30–35.
Strohecker AM, Joshi S, Possemato R, Abraham RT, Sabatini DM, White E . Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel autophagy regulator by high content shRNA screening. Oncogene 2015; 34: 5662–5676.
Willis A, Jung EJ, Wakefield T, Chen X . Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 2004; 23: 2330–2338.
Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E . The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64: 2627–2633.
Bengoechea-Alonso MT, Ericsson J . SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol 2007; 19: 215–222.
Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012; 148: 244–258.
Yahagi N, Shimano H, Matsuzaka T, Najima Y, Sekiya M, Nakagawa Y et al. p53 Activation in adipocytes of obese mice. J Biol Chem 2003; 278: 25395–25400.
Denko NC . Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer 2008; 8: 705–713.
Mayers JR, Vander Heiden MG . Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci 2015; 40: 130–140.
Casciari JJ, Sotirchos SV, Sutherland RM . Variations in tumor cell growth rates and metabolism with oxygen concentration, glucose concentration, and extracellular pH. J Cell Physiol 1992; 151: 386–394.
Maddocks OD, Vousden KH . Metabolic regulation by p53. J Mol Med 2011; 89: 237–245.
Kim HR, Roe JS, Lee JE, Cho EJ, Youn HD . p53 regulates glucose metabolism by miR-34a. Biochem Biophys Res Commun 2013; 437: 225–231.
Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013; 493: 542–546.
Rinn JL, Huarte M . To repress or not to repress: this is the guardian's question. Trends Cell Biol 2011; 21: 344–353.
Ros S, Schulze A . Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab 2013; 1: 8.
Chesney J, Clark J, Klarer AC, Imbert-Fernandez Y, Lane AN, Telang S . Fructose-2,6-bisphosphate synthesis by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 2014; 5: 6670–6686.
Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 2007; 67: 3043–3053.
Laplante M, Sabatini DM . An emerging role of mTOR in lipid biosynthesis. Curr Biol 2009; 19: R1046–R1052.
Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol 2007; 6: 10.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282: 1497–1501.
Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D . Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 1997; 15: 871–875.
Van Schaftingen E, Lederer B, Bartrons R, Hers HG . A kinetic study of pyrophosphate: fructose-6-phosphate phosphotransferase from potato tubers. Application to a microassay of fructose 2,6-bisphosphate. Eur J Biochem 1982; 129: 191–195.
Buescher JM, Moco S, Sauer U, Zamboni N . Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem 2010; 82: 4403–4412.
Ruhl M, Rupp B, Noh K, Wiechert W, Sauer U, Zamboni N . Collisional fragmentation of central carbon metabolites in LC-MS/MS increases precision of (1)(3)C metabolic flux analysis. Biotechnol Bioeng 2012; 109: 763–771.
Swinnen JV, Heemers H, Deboel L, Foufelle F, Heyns W, Verhoeven G . Stimulation of tumor-associated fatty acid synthase expression by growth factor activation of the sterol regulatory element-binding protein pathway. Oncogene 2000; 19: 5173–5181.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D et al. DAVID Bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007; 35: W169–W175.
Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 2006; 66: 10292–10301.
Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res 2008; 14: 7397–7404.
Goswami CP, Nakshatri H . PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinform 2013; 3: 22.
Acknowledgements
We thank C. Esnault (LRI) for advice on ChIP and H. Miess for help with Seahorse Analysis, C. Watkins and J. Bee (LRI, BRU) for help with animal experimentation, F. Lassailly, P. Johnson and T. Snoeks (In vivo imaging facility, LRI) for assistance with in vivo imaging and the LRI research services for technical support. We also thank C. Ade and Barbara Bauer (Theodor-Boveri-Institute, Würzburg) for help with RNAseq analysis and histology, Beatrice Dankworth for help with cell line generation and W. Schmitz for insightful discussions. Cancer Research UK, the German Cancer Aid (grant 111917), the German Research Foundation (FOR 2314), and the CRUK-EPSRC Imaging Centre in Cambridge and Manchester (grant 16465) supported this work.
Author contributions
SR and AS conceived the project and wrote the manuscript. SR, JF, IK, CD, AH, SD, BG and SB performed experiments and analysis of results. RM and SW performed the bioinformatic analysis. AB, KMB, NZ and MHR contributed to the study design and data analysis. All authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Ros, S., Flöter, J., Kaymak, I. et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene 36, 3287–3299 (2017). https://doi.org/10.1038/onc.2016.477
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.477
This article is cited by
-
Cuproptosis: p53-regulated metabolic cell death?
Cell Death & Differentiation (2023)
-
Hypoxia-mediated promotion of glucose metabolism in non-small cell lung cancer correlates with activation of the EZH2/FBXL7/PFKFB4 axis
Cell Death & Disease (2023)
-
Differential roles of highly expressed PFKFB4 in colon adenocarcinoma patients
Scientific Reports (2023)
-
The role of ROS in tumour development and progression
Nature Reviews Cancer (2022)
-
Therapeutic and preventive effects of exercise training on metabolic regulators/markers in mouse colorectal cancer cells
Sport Sciences for Health (2022)