Abstract
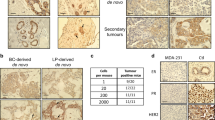
Screening for oncogenes has mostly been performed by in vitro transformation assays. However, some oncogenes might not exhibit their transforming activities in vitro unless putative essential factors from in vivo microenvironments are adequately supplied. Here, we have developed an in vivo screening system that evaluates the tumorigenicity of target genes. This system uses a retroviral high-efficiency gene transfer technique, a large collection of human cDNA clones corresponding to ~70% of human genes and a luciferase-expressing immortalized mouse mammary epithelial cell line (NMuMG-luc). From 845 genes that were highly expressed in human breast cancer cell lines, we focused on 205 genes encoding membrane proteins and/or kinases as that had the greater possibility of being oncogenes or drug targets. The 205 genes were divided into five subgroups, each containing 34–43 genes, and then introduced them into NMuMG-luc cells. These cells were subcutaneously injected into nude mice and monitored for tumor development by in vivo imaging. Tumors were observed in three subgroups. Using DNA microarray analyses and individual tumorigenic assays, we found that three genes, ADORA2B, PRKACB and LPAR3, were tumorigenic. ADORA2B and LPAR3 encode G-protein-coupled receptors and PRKACB encodes a protein kinase A catalytic subunit. Cells overexpressing ADORA2B, LPAR3 or PRKACB did not show transforming phenotypes in vitro, suggesting that transformation by these genes requires in vivo microenvironments. In addition, several clinical data sets, including one for breast cancer, showed that the expression of these genes correlated with lower overall survival rate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Matsui A, Ihara T, Suda H, Mikami H, Semba K . Gene amplification: mechanisms and involvement in cancer. Biomol Concepts 2013; 4: 567–582.
Saito M, Kato Y, Ito E, Fujimoto J, Ishikawa K, Doi A et al. Expression screening of 17q12–21 amplicon reveals GRB7 as an ERBB2-dependent oncogene. FEBS Lett 2012; 586: 1708–1714.
Doi A, Ishikawa K, Shibata N, Ito E, Fujimoto J, Yamamoto M et al. Enhanced expression of retinoic acid receptor alpha (RARA) induces epithelial-to-mesenchymal transition and disruption of mammary acinar structures. Mol Oncol 2015; 9: 355–364.
Polanska UM, Orimo A . Carcinoma-associated fibroblasts: non-neoplastic tumour-promoting mesenchymal cells. J Cell Physiol 2013; 228: 1651–1657.
Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature 1993; 362: 841–844.
Fasano O, Birnbaum D, Edlund L, Fogh J, Wigler M . New human transforming genes detected by a tumorigenicity assay. Mol Cell Biol 1984; 4: 1695–1705.
Young D, Waitches G, Birchmeier C, Fasano O, Wigler M . Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell 1986; 45: 711–719.
O' Hayre M, Degese MS, Gutkind JS . Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol 2014; 27: 126–135.
Goshima N, Kawanura Y, Fukumoto A, Miura A, Homma R, Satoh R et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Methods 2008; 5: 1011–1017.
Hynes NE, Jaggi R, Kozma SC, Ball R, Muellener D, Wetherall NT et al. New acceptor cell for transfected genomic DNA: oncogene transfer into a mouse mammary epithelial cell line. Mol Cell Biol 1985; 5: 268–272.
Ito E, Honma R, Yanagisawa Y, Imai J, Azuma S, Oyama T et al. Novel clusters of highly expressed genes accompany genomic amplification in breast cancers. FEBS Lett 2007; 581: 3909–3914.
van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999–2009.
Dorsam RT, Gutkind JS . G-protein-coupled receptors and cancer. Nat Rev Cancer 2007; 7: 79–94.
Parma J, Duprez L, Van Sande J, Cochaux P, Gervy C, Mockel J et al. Somatic mutations in the thyrotropin receptor gene cause hyperfunctioning thyroid adenomas. Nature 1993; 365: 649–651.
Feigin ME . Harnessing the genome for characterization of G-protein coupled receptors in cancer pathogenesis. FEBS J 2013; 280: 4729–4738.
Feoktistov I, Biaggioni I . Adenosine A2B receptors. Pharmacol Rev 1997; 49: 381–402.
Fredholm BB . Adenosine, an endogenous distress signal, modulates tissue damage and repair. Cell Death Differ 2007; 14: 1315–1323.
Antonioli L, Blandizzi C, Pacher P, Haskó G . Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer 2013; 13: 842–857.
Desmet CJ, Gallenne T, Prieur A, Reyal F, Visser NL, Wittner BS et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci USA 2013; 110: 5139–5144.
Ntantie E, Gonyo P, Lorimer EL, Hauser AD, Schuld N, McAllister D et al. An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci Signal 2013; 6: ra39.
Stagg J, Divisekera U, Mclaughlin N, Sharkey J, Pommey S, Denoyer D et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Nat Acad Sci USA 2010; 107: 1547–1552.
Mutoh T, Rivera R, Chun J . Insights into the pharmacological relevance of lysophospholipid receptors. Br J Pharmacol 2012; 165: 829–844.
Millis GB, Moolenaar WH . The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer 2003; 3: 582–591.
Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W, Zhang F et al. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell 2009; 15: 539–550.
Naviglio S, Caraglia M, Abbruzzese A, Chiosi E, Di Gesto D, Marra M et al. Protein kinase A as a biological target in cancer therapy. Expert Opin Ther Targets 2009; 13: 83–92.
Correa R, Salpea P, Stratakis CA . Carney complex: an update. Eur J Endocrinol 2015; 173: M85–M97.
Kirschner LS, Kusewitt DF, Matyakhina L, Towns WH II, Carney JA, Westphal H et al. A mouse model for the Carney complex tumor syndrome develops neoplasia in cyclic AMP-responsive tissues. Cancer Res 2005; 65: 4506–4514.
Griffin KJ, Kirschner LS, Matyakhina L, Stergiopoulos SG, Robinson-White A, Lenherr SM et al. A transgenic mouse bearing an antisense construct of regulatory subunit type 1A of protein kinase A develops endocrine and other tumours: comparison with Carney complex and other PRKAR1A induced lesions. J Med Genet 2004; 41: 923–931.
Beristain AG, Molyneux SD, Joshi PA, Pomroy NC, Di Grappa MA, Chang MC et al. PKA signaling drives mammary tumorigenesis through Src. Oncogene 2015; 34: 1160–1173.
Padmanabhan A, Li X, Bieberich CJ . Protein kinase A regulates MYC protein through transcriptional and post-translational mechanisms in a catalytic subunit isoform-specific manner. J Biol Chem 2013; 288: 14158–14169.
Acknowledgements
We thank our laboratory members, especially Kumiko Semba for secretarial assistance and Sakura Azuma-Tamamura for technical assistance. This work was conducted as part of the Fukushima Translational Research Project and supported, in part, by JSPS KAKENHI Grant Number 23241064, and a Grant-in-Aid for Scientific Research (A).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Ihara, T., Hosokawa, Y., Kumazawa, K. et al. An in vivo screening system to identify tumorigenic genes. Oncogene 36, 2023–2029 (2017). https://doi.org/10.1038/onc.2016.351
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.351
This article is cited by
-
Protective potential of miR-146a-5p and its underlying molecular mechanism in diverse cancers: a comprehensive meta-analysis and bioinformatics analysis
Cancer Cell International (2019)
-
A method of producing genetically manipulated mouse mammary gland
Breast Cancer Research (2019)
-
Integration of multiple types of genetic markers for neuroblastoma may contribute to improved prediction of the overall survival
Biology Direct (2018)