Abstract
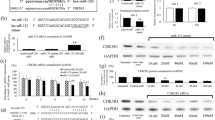
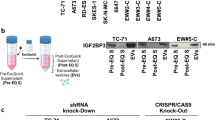
Sarcomas are mesenchymal tumors characterized by blocked differentiation process. In Ewing sarcoma (EWS) both CD99 and EWS-FLI1 concur to oncogenesis and inhibition of differentiation. Here, we demonstrate that uncoupling CD99 from EWS-FLI1 by silencing the former, nuclear factor-κB (NF-κB) signaling is inhibited and the neural differentiation program is re-established. NF-κB inhibition passes through miR-34a-mediated repression of Notch pathway. CD99 counteracts EWS-FLI1 in controlling NF-κB signaling through the miR-34a, which is increased and secreted into exosomes released by CD99-silenced EWS cells. Delivery of exosomes from CD99-silenced cells was sufficient to induce neural differentiation in recipient EWS cells through miR-34a inhibition of Notch-NF-κB signaling. Notably, even the partial delivery of CD99 small interfering RNA may have a broad effect on the entire tumor cell population owing to the spread operated by their miR-34a-enriched exosomes, a feature opening to a new therapeutic option.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. J Clin Oncol 2012; 30: 4148–4154.
Magnan H, Goodbody CM, Riedel E, Pratilas CA, Wexler LH, Chou AJ . Ifosfamide dose-intensification for patients with metastatic Ewing sarcoma. Pediatr Blood Cancer 2015; 62: 594–597.
Ginsberg JP, Goodman P, Leisenring W, Ness KK, Meyers PA, Wolden SL et al. Long-term survivors of childhood Ewing sarcoma: report from the childhood cancer survivor study. J Natl Cancer Inst 2010; 102: 1272–1283.
Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992; 359: 162–165.
Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT . A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet 1994; 6: 146–151.
Lessnick SL, Ladanyi M . Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev Pathol 2012; 7: 145–159.
Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 2008; 68: 2176–2185.
Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O . Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007; 11: 421–429.
Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES . Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene 1995; 11: 1049–1054.
Maksimenko A, Malvy C . Oncogene-targeted antisense oligonucleotides for the treatment of Ewing sarcoma. Expert Opin Ther Targets 2005; 9: 825–830.
Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014; 4: 1342–1353.
Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014; 4: 1326–1341.
Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S et al. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014; 10: e1004475.
Armengol G, Tarkkanen M, Virolainen M, Forus A, Valle J, Bohling T et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridization. Br J Cancer 1997; 75: 1403–1409.
Lopez-Guerrero JA, Pellin A, Noguera R, Carda C, Llombart-Bosch A . Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest 2001; 81: 803–814.
Huang HY, Illei PB, Zhao Z, Mazumdar M, Huvos AG, Healey JH et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol 2005; 23: 548–558.
Lessnick SL, Dacwag CS, Golub TR . The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 2002; 1: 393–401.
Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ . The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem 1997; 272: 30822–30827.
Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest 2010; 120: 668–680.
Gelin C, Aubrit F, Phalipon A, Raynal B, Cole S, Kaczorek M et al. The E2 antigen, a 32 kd glycoprotein involved in T-cell adhesion processes, is the MIC2 gene product. EMBO J 1989; 8: 3253–3259.
Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA . CD99 plays a major role in the migration of monocytes through endothelial junctions. Nat Immunol 2002; 3: 143–150.
Bernard G, Breittmayer JP, de Matteis M, Trampont P, Hofman P, Senik A et al. Apoptosis of immature thymocytes mediated by E2/CD99. J Immunol 1997; 158: 2543–2550.
Alberti I, Bernard G, Rouquette-Jazdanian AK, Pelassy C, Pourtein M, Aussel C et al. CD99 isoforms expression dictates T cell functional outcomes. FASEB J 2002; 16: 1946–1948.
Miyagawa Y, Okita H, Nakaijima H, Horiuchi Y, Sato B, Taguchi T et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing's family tumor-like phenotypes to human mesenchymal progenitor cells. Mol Cell Biol 2008; 28: 2125–2137.
Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ . EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors. Cancer Res 2005; 65: 4633–4644.
Amaral AT, Manara MC, Berghuis D, Ordonez JL, Biscuola M, Lopez-Garcia MA et al. Characterization of human mesenchymal stem cells from ewing sarcoma patients. Pathogenetic implications. PLoS One 2014; 9: e85814.
Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O . MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene 2013; 32: 3915–3921.
Charytonowicz E, Terry M, Coakley K, Telis L, Remotti F, Cordon-Cardo C et al. PPARgamma agonists enhance ET-743-induced adipogenic differentiation in a transgenic mouse model of myxoid round cell liposarcoma. J Clin Invest 2012; 122: 886–898.
Hayden MS, Ghosh S . Shared principles in NF-kappaB signaling. Cell 2008; 132: 344–362.
Viatour P, Merville MP, Bours V, Chariot A . Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci 2005; 30: 43–52.
Rorie CJ, Thomas VD, Chen P, Pierce HH, O'Bryan JP, Weissman BE . The Ews/Fli-1 fusion gene switches the differentiation program of neuroblastomas to Ewing sarcoma/peripheral primitive neuroectodermal tumors. Cancer Res 2004; 64: 1266–1277.
Teitell MA, Thompson AD, Sorensen PH, Shimada H, Triche TJ, Denny CT . EWS/ETS fusion genes induce epithelial and neuroectodermal differentiation in NIH 3T3 fibroblasts. Lab Invest 1999; 79: 1535–1543.
Gonzalez I, Vicent S, de Alava E, Lecanda F . EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. J Mol Med (Berl) 2007; 85: 1015–1029.
Hayden MS, Ghosh S . Regulation of NF-kappaB by TNF family cytokines. Semin Immunol 2014; 26: 253–266.
Sasaki CY, Barberi TJ, Ghosh P, Longo DL . Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J Biol Chem 2005; 280: 34538–34547.
Staudt LM . Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol 2010; 2: a000109.
Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH . Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res 2006; 66: 2778–2784.
Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH . Inhibition of nuclear factor kappab activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer 2006; 118: 1930–1936.
Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S et al. miR-34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol 2012; 226: 796–805.
Marino MT, Grilli A, Baricordi C, Manara MC, Ventura S, Pinca RS et al. Prognostic significance of miR-34a in Ewing sarcoma is associated with cyclin D1 and ki-67 expression. Ann Oncol 2014; 25: 2080–2086.
Prasad S, Ravindran J, Aggarwal BB . NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem 2010; 336: 25–37.
Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Lafarga M, Berciano MT, Aldaz B et al. Blockade of the NFkappaB pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene 2011; 30: 3537–3548.
Sabolek M, Herborg A, Schwarz J, Storch A . Dexamethasone blocks astroglial differentiation from neural precursor cells. NeuroReport 2006; 17: 1719–1723.
Ozeki A, Suzuki K, Suzuki M, Ozawa H, Yamashita S . Acceleration of astrocytic differentiation in neural stem cells surviving X-irradiation. NeuroReport 2012; 23: 290–293.
De Molfetta GA, Luciola Zanette D, Alexandre Panepucci R, Dos Santos AR, da Silva WA Jr, Antonio Zago M . Role of NFKB2 on the early myeloid differentiation of CD34+ hematopoietic stem/progenitor cells. Differentiation 2010; 80: 195–203.
Vaira S, Johnson T, Hirbe AC, Alhawagri M, Anwisye I, Sammut B et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc Natl Acad Sci USA 2008; 105: 3897–3902.
Ahmed AA, Sherman AK, Pawel BR . Expression of therapeutic targets in Ewing sarcoma family tumors. Hum Pathol 2012; 43: 1077–1083.
Javelaud D, Wietzerbin J, Delattre O, Besancon F . Induction of p21Waf1/Cip1 by TNFalpha requires NF-kappaB activity and antagonizes apoptosis in Ewing tumor cells. Oncogene 2000; 19: 61–68.
Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem 2006; 281: 30373–30382.
Javelaud D, Poupon MF, Wietzerbin J, Besancon F . Inhibition of constitutive NF-kappa B activity suppresses tumorigenicity of Ewing sarcoma EW7 cells. Int J Cancer 2002; 98: 193–198.
Sonnemann J, Dreyer L, Hartwig M, Palani CD, Hong le TT, Klier U et al. Histone deacetylase inhibitors induce cell death and enhance the apoptosis-inducing activity of TRAIL in Ewing's sarcoma cells. J Cancer Res Clin Oncol 2007; 133: 847–858.
Sonnemann J, Palani CD, Wittig S, Becker S, Eichhorn F, Voigt A et al. Anticancer effects of the p53 activator nutlin-3 in Ewing's sarcoma cells. Eur J Cancer 2011; 47: 1432–1441.
Lagirand-Cantaloube J, Laud K, Lilienbaum A, Tirode F, Delattre O, Auclair C et al. EWS-FLI1 inhibits TNFalpha-induced NFkappaB-dependent transcription in Ewing sarcoma cells. Biochem Biophys Res Commun 2010; 399: 705–710.
von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One 2011; 6: e19305.
Widera D, Mikenberg I, Kaltschmidt B, Kaltschmidt C . Potential role of NF-kappaB in adult neural stem cells: the underrated steersman? Int J Dev Neurosci 2006; 24: 91–102.
Yang C, Atkinson SP, Vilella F, Lloret M, Armstrong L, Mann DA et al. Opposing putative roles for canonical and noncanonical NFkappaB signaling on the survival, proliferation, and differentiation potential of human embryonic stem cells. Stem Cells 2010; 28: 1970–1980.
Hess K, Ushmorov A, Fiedler J, Brenner RE, Wirth T . TNFalpha promotes osteogenic differentiation of human mesenchymal stem cells by triggering the NF-kappaB signaling pathway. Bone 2009; 45: 367–376.
Cho HH, Shin KK, Kim YJ, Song JS, Kim JM, Bae YC et al. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J Cell Physiol 2010; 223: 168–177.
Osipo C, Golde TE, Osborne BA, Miele LA . Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest 2008; 88: 11–17.
Ban J, Bennani-Baiti IM, Kauer M, Schaefer KL, Poremba C, Jug G et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma. Cancer Res 2008; 68: 7100–7109.
Bennani-Baiti IM, Aryee DN, Ban J, Machado I, Kauer M, Muhlbacher K et al. Notch signalling is off and is uncoupled from HES1 expression in Ewing's sarcoma. J Pathol 2011; 225: 353–363.
Wang Y, Chan SL, Miele L, Yao PJ, Mackes J, Ingram DK et al. Involvement of Notch signaling in hippocampal synaptic plasticity. Proc Natl Acad Sci USA 2004; 101: 9458–9462.
Baliko F, Bright T, Poon R, Cohen B, Egan SE, Alman BA . Inhibition of notch signaling induces neural differentiation in Ewing sarcoma. Am J Pathol 2007; 170: 1686–1694.
Pang RT, Leung CO, Lee CL, Lam KK, Ye TM, Chiu PC et al. MicroRNA-34a is a tumor suppressor in choriocarcinoma via regulation of Delta-like1. BMC Cancer 2013; 13: 25.
Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA . Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ 2015; 22: 34–45.
Pata S, Otahal P, Brdicka T, Laopajon W, Mahasongkram K, Kasinrerk W . Association of CD99 short and long forms with MHC class I, MHC class II and tetraspanin CD81 and recruitment into immunological synapses. BMC Res Notes 2011; 4: 293.
Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res 2007; 13: 2429–2440.
Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res 2012; 18: 6260–6270.
Garofalo C, Mancarella C, Grilli A, Manara MC, Astolfi A, Marino MT et al. Identification of common and distinctive mechanisms of resistance to different anti-IGF-IR agents in Ewing's sarcoma. Mol Endocrinol 2012; 26: 1603–1616.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ . Normalization panels for the reliable quantification of circulating microRNAs by RT–qPCR. FASEB J 2015; 29: 3853–3862.
McDermott AM, Kerin MJ, Miller N . Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One 2013; 8: e83718.
Acknowledgements
We are indebted to Cristina Ghinelli for editing the manuscript. We wish to thank Dr Gianfranco Mattia for the assessment of immunofluorescence analysis on the exosome fusion. We are also grateful to Prof. Alain Israel and Dr Johannes Schmid for sharing NF-κB reporter and plasmids. This work was supported by grants from the Italian Association for Cancer Research (AIRC project: IG2013_14049 to KS; IG2012_13247 to AC), Ministry of Education, Research and Universities (FIRB project: RBAP11884 M_005 to KS), Ministry of Health (PROVABES project: PER-2011-2353839 to PP and KS), Austrian Science Fund (project: P24708-B21 and PROVABES project: I-1225-B19 to HK). Selena Ventura is in receipt of the fellowship ‘Guglielmina Lucatello e Gino Mazzega’ granted by Fondazione Italiana per la Ricerca sul Cancro (FIRC project code: 13811).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Ventura, S., Aryee, D., Felicetti, F. et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-κB signaling. Oncogene 35, 3944–3954 (2016). https://doi.org/10.1038/onc.2015.463
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.463
This article is cited by
-
The basic characteristics of extracellular vesicles and their potential application in bone sarcomas
Journal of Nanobiotechnology (2021)
-
miR-34a induces immunosuppression in colorectal carcinoma through modulating a SIRT1/NF-κB/B7-H3/TNF-α axis
Cancer Immunology, Immunotherapy (2021)
-
Exosomes from CD99-deprived Ewing sarcoma cells reverse tumor malignancy by inhibiting cell migration and promoting neural differentiation
Cell Death & Disease (2019)
-
miRNA signatures in childhood sarcomas and their clinical implications
Clinical and Translational Oncology (2019)
-
Clofarabine inhibits Ewing sarcoma growth through a novel molecular mechanism involving direct binding to CD99
Oncogene (2018)