Abstract
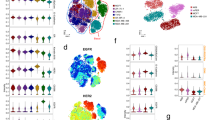
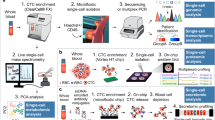
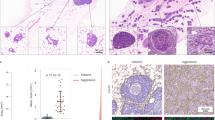
Tumor heterogeneity and the presence of drug-sensitive and refractory populations within the same tumor are almost never assessed in the drug discovery pipeline. Such incomplete assessment of drugs arising from spatial and temporal tumor cell heterogeneity reflects on their failure in the clinic and considerable wasted costs in the drug discovery pipeline. Here we report the derivation of a flow cytometry-based tumor deconstruction platform for resolution of at least 18 discrete tumor cell fractions. This is achieved through concurrent identification, quantification and analysis of components of cancer stem cell hierarchies, genetically instable clones and differentially cycling populations within a tumor. We also demonstrate such resolution of the tumor cytotype to be a potential value addition in drug screening through definitive cell target identification. Additionally, this real-time definition of intra-tumor heterogeneity provides a convenient, incisive and analytical tool for predicting drug efficacies through profiling perturbations within discrete tumor cell subsets in response to different drugs and candidates. Consequently, possible applications in informed therapeutic monitoring and drug repositioning in personalized cancer therapy would complement rational design of new candidates besides achieving a re-evaluation of existing drugs to derive non-obvious combinations that hold better chances of achieving remission.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wang B, Rosano JM, Cheheltani R, Achary MP, Kiani MF . Towards a targeted multi-drug delivery approach to improve therapeutic efficacy in breast cancer. Expert Opin Drug Deliv 2010; 7: 1159–1173.
Wani AA, Sharma N, Shouche YS, Bapat SA . Nuclear-mitochondrial genomic profiling reveals a pattern of evolution in epithelial ovarian tumor stem cells. Oncogene 2006; 25: 6336–6344.
Navin NE, Hicks J . Tracing the tumor lineage. Mol Oncol 2010; 4: 267–283.
Marusyk A, Polyak K . Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 2010; 1805: 105–117.
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Li B, Senbabaoglu Y, Peng W, Yang ML, Xu J, Li JZ . Genomic estimates of aneuploid content in glioblastoma multiforme and improved classification. Clin Cancer Res 2012; 18: 5595–5605.
Magee JA, Piskounova E, Morrison SJ . Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 2012; 3: 283–296.
Schoenborn JR, Nelson P, Fang M . Genomic profiling defines subtypes of prostate cancer with the potential for therapeutic stratification. Clin Cancer Res 2013; 15: 4058–4066.
Fisher R, Pusztai L, Swanton C . Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer 2013; 108: 479–485.
Visvader JE, Lindeman GJ . Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10: 717–728.
Cree IA, Glaysher S, Harvey AL . Efficacy of anti-cancer agents in cell lines versus human primary tumour tissue. Curr Opin Pharmacol 2010; 4: 375–379.
Bapat SA, Mali AM, Koppikar CB, Kurrey NK . Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res 2005; 65: 3025–3029.
Kusumbe AP, Bapat SA . Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res 2009; 69: 9245–1953.
Kusumbe AP, Mali AM, Bapat SA . CD133-expressing stem cells associated with ovarian metastases establish an endothelial hierarchy and contribute to tumor vasculature. Stem Cells 2009; 27: 498–508.
Bouchet BP, Bertholon J, Falette N, Audoynaud C, Lamblot C, Puisieux A et al. Paclitaxel resistance in untransformed human mammary epithelial cells is associated with an aneuploidy-prone phenotype. Br J Cancer 2007; 97: 1218–1224.
Bian M, Fu J, Yan Y, Chen Q, Yang C, Shi Q et al. Short exposure to paclitaxel induces multipolar spindle formation and aneuploidy through promotion of acentrosomal pole assembly. Sci China Life Sci 2010; 53: 1322–1329.
Kunos CA, Ferris G, Pyatka N, Pink J, Radivoyevitch T . Deoxynucleoside salvage facilitates DNA repair during ribonucleotide reductase blockade in human cervical cancers. Radiat Res 2011; 176: 425–433.
Chiang Y, Davis RG, Vishwanatha JK . Altered expression of annexin II in human B-cell lymphoma cell lines. Biochim Biophys Acta 1996; 1313: 295–301.
Ye NS, Zhang RL, Zhao YF, Feng X, Wang YM, Luo GA . Effect of 5-azacytidine on the protein expression of porcine bone marrow mesenchymal stem cells in vitro. Genomics Proteomics Bioinformatics 2006; 4: 18–25.
Niu D, Sui J, Zhang J, Feng H, Chen WN . iTRAQ-coupled 2-D LC-MS/MS analysis of protein profile associated with HBV-modulated DNA methylation. Proteomics 2009; 9: 3856–3868.
Diaz-Cano SJ . Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. Int J Mol Sci 2012; 13: 1951–2011.
Cavanagh BL, Walker T, Norazit A, Meedeniya AC . Thymidine analogues for tracking DNA synthesis. Molecules 2011; 9: 7980–7993.
Shan-Cheng R, Min Q, Ying-Hao S . Investigating intratumour heterogeneity by single-cell sequencing. Asian J Androl 2013; 15: 729–734.
Junttila MR, de Sauvage FJ . Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013; 501: 346–354.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 10: 883–892.
Al-Lazikani B, Banerji U, Workman P . Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 2012; 7: 679–692.
Natrajan R, Wilkerson P . From integrative genomics to therapeutic targets. Cancer Res 2013; 73: 3483–3488.
McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med 2010; 16: 483–489.
Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 2013; 3: 1416–1429.
Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM . Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol 2013; 10: 483–487.
Lin D, Xue H, Wang Y, Wu R, Watahiki A, Dong X et al. Next generation patient-derived prostate cancer xenograft models. Asian J Androl 2014; 16: 407–412.
Cleary AS, Leonard TL, Gestl SA, Gunther EJ . Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014; 7494: 113–117.
Hu Y, Smyth GK . ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009; 347: 70–78.
Acknowledgements
We appreciate the efforts of Dr Mrunalini Moghe (Deenanath Mangeshkar Hospital, Pune) and Dr Anjali Kusumbe in karyotyping, Mr Swapnil Kamble in cryosectioning and thank Dr Shona Nag (Jehangir Hospital, Pune) for providing gemcitabine. This research is supported by NCCS intra-mural grants to SAB.
Author Contributions
RRN performed experiments, analyzed data, assisted manuscript drafting and carried out reviewer suggested revisions; AKS performed epigenetic drug-associated experiments, analyzed data and assisted manuscript drafting; AMM assisted with in vivo experiments; MFK analyzed data for derivation of checker board representation of drug responses; and SAB conceived the project, planned and finalized experimental design, analyzed data, drafted and edited the manuscript and its revision.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Following patents have been filed relating to the findings of this manuscript, 173/MUM/2014, PCT/IB2015/050358, and Indian provisional patent application 2980/MUM/2014.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Naik, R., Singh, A., Mali, A. et al. A tumor deconstruction platform identifies definitive end points in the evaluation of drug responses. Oncogene 35, 727–737 (2016). https://doi.org/10.1038/onc.2015.130
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.130
This article is cited by
-
Elucidation of molecular and functional heterogeneity through differential expression network analyses of discrete tumor subsets
Scientific Reports (2016)
-
Bioluminescence Microscopy as a Method to Measure Single Cell Androgen Receptor Activity Heterogeneous Responses to Antiandrogens
Scientific Reports (2016)