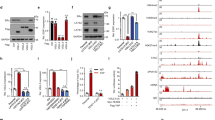
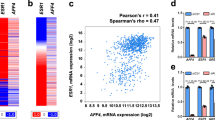
Abstract
Deleted in Breast Cancer 1 (DBC1), a negative regulator of deacetylase SIRT1, has been shown to act as an estrogen receptor α (ER) coactivator that has a key role in ER transcription complex assembly and estrogen-dependent breast cancer cell proliferation. However, little is known about its physiological role and mechanism of action in ER-negative breast cancer cells. Here we report that DBC1 functions as a coactivator for the oncogenic ETS transcription factor PEA3 to promote ER-negative breast cancer progression. DBC1 is required for the expression of PEA3 target genes and for recruitment of PEA3 and RNA polymerase II to PEA3 target promoters. We also demonstrated that acetylation of PEA3 stimulates its DNA binding and association with DBC1 by disrupting the intramolecular interaction of PEA3. The molecular mechanism underlying DBC1 function in PEA3-mediated transcription involves inhibition of SIRT1 interaction with PEA3 and of SIRT1-mediated deacetylation of PEA3. Moreover, DBC1 depletion inhibited the tumorigenic properties of ER-negative breast cancer cells in vitro and in vivo. Importantly, increased DBC1 expression correlated with shorter relapse-free survival of ER-negative breast cancer patients. Our results firmly established DBC1 as a critical coactivator of PEA3 and as a key player in PEA3-mediated breast cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–10874.
Reis-Filho JS, Pusztai L . Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet 2011; 378: 1812–1823.
Barcellos-Hoff MH . Does microenvironment contribute to the etiology of estrogen receptor-negative breast cancer? Clin Cancer Res 2013; 19: 541–548.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Sharrocks AD . The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2: 827–837.
Oh S, Shin S, Janknecht R . ETV1 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012; 1826: 1–12.
Myers E, Hill AD, Kelly G, McDermott EW, O'Higgins NJ, Young LS . A positive role for PEA3 in HER2-mediated breast tumour progression. Br J Cancer 2006; 95: 1404–1409.
Benz CC, O'Hagan RC, Richter B, Scott GK, Chang CH, Xiong X et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene 1997; 15: 1513–1525.
O'Hagan RC, Tozer RG, Symons M, McCormick F, Hassell JA . The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene 1996; 13: 1323–1333.
Guo B, Panagiotaki N, Warwood S, Sharrocks AD . Dynamic modification of the ETS transcription factor PEA3 by sumoylation and p300-mediated acetylation. Nucleic Acids Res 2011; 39: 6403–6413.
Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH . Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor α activity and co-activator synergy. Nucleic Acids Res 2011; 39: 6932–6943.
Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W . Negative regulation of the deacetylase SIRT1 by DBC1. Nature 2008; 451: 587–590.
Kim JE, Chen J, Lou Z . DBC1 is a negative regulator of SIRT1. Nature 2008; 451: 583–586.
Fu J, Jiang J, Li J, Wang S, Shi G, Feng Q et al. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J Biol Chem 2009; 284: 6832–6840.
Chini CC, Escande C, Nin V, Chini EN . HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem 2010; 285: 40830–40837.
Gu S, Chen L, Hong Q, Yan T, Zhuang Z, Wang Q et al. PEA3 activates CXCR4 transcription in MDA-MB-231 and MCF7 breast cancer cells. Acta Biochim Biophys Sin 2011; 43: 771–778.
Sun Y, Wenger L, Brinckerhoff CE, Misra RR, Cheung HS . Basic calcium phosphate crystals induce matrix metalloproteinase-1 through the Ras/mitogen-activated protein kinase/c-Fos/AP-1/metalloproteinase 1 pathway. Involvement of transcription factor binding sites AP-1 and PEA-3. J Biol Chem 2002; 277: 1544–1552.
Wollenick K, Hu J, Kristiansen G, Schraml P, Rehrauer H, Berchner-Pfannschmidt U et al. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res 2012; 40: 1928–1943.
Bohn OL, Nasir I, Brufsky A, Tseng GC, Bhargava R, MacManus K et al. Biomarker profile in breast carcinomas presenting with bone metastasis. Int J Clin Exp Pathol 2009; 3: 139–146.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA . The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol 2001; 11: 1739–1748.
Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 2009; 36: 153–163.
Greenall A, Willingham N, Cheung E, Boam DS, Sharrocks AD . DNA binding by the ETS-domain transcription factor PEA3 is regulated by intramolecular and intermolecular protein.protein interactions. J Biol Chem 2001; 276: 16207–16215.
Bojovic BB, Hassell JA . The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding. J Biol Chem 2001; 276: 4509–4521.
Gu W, Roeder RG . Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997; 90: 595–606.
Guo B, Sharrocks AD . Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol Cell Biol 2009; 29: 3204–3218.
Baert JL, Monte D, Musgrove EA, Albagli O, Sutherland RL, de Launoit Y . Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int J Cancer 1997; 70: 590–597.
Goel A, Janknecht R . Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol 2003; 23: 6243–6254.
Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol 2011; 42: 204–213.
Hiraike H, Wada-Hiraike O, Nakagawa S, Saji S, Maeda D, Miyamoto Y et al. Expression of DBC1 is associated with nuclear grade and HER2 expression in breast cancer. Exp Ther Med 2011; 2: 1105–1109.
Eck SM, Cote AL, Winkelman WD, Brinckerhoff CE . CXCR4 and matrix metalloproteinase-1 are elevated in breast carcinoma-associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol Cancer Res 2009; 7: 1033–1044.
Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A . Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther 2013; 6: 1347–1361.
Seo WY, Jeong BC, Yu EJ, Kim HJ, Kim SH, Lim JE et al. CCAR1 promotes chromatin loading of androgen receptor (AR) transcription complex by stabilizing the association between AR and GATA2. Nucleic Acids Res 2013; 41: 8526–8536.
Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR . CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell 2008; 31: 510–519.
Yu EJ, Kim SH, Kim MJ, Seo WY, Song KA, Kang MS et al. SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. Oncogene 2013; 32: 4160–4168.
Choi YL, Oh E, Park S, Kim Y, Park YH, Song K et al. Triple-negative, basal-like, and quintuple-negative breast cancers: better prediction model for survival. BMC Cancer 2010; 10: 507.
Kim SH, Kim JH, Yu EJ, Lee KW, Park CK . The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol Histopathol 2012; 27: 49–58.
Acknowledgements
We thank Jason W Chin (MRC Laboratory for Molecular Biology, Cambridge, UK) for providing vectors and Yoon-La Choi (Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea) for providing the tissue microarray slides of primary invasive breast carcinoma specimens. This work was supported by Samsung Biomedical Research Institute (SBRI) grant (SMX1132501) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A1A2059697) to JHK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, H., Kim, SH., Yu, E. et al. A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer. Oncogene 34, 4500–4508 (2015). https://doi.org/10.1038/onc.2014.381
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.381
This article is cited by
-
DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer
Cell Death & Disease (2018)
-
DBC1 promotes castration-resistant prostate cancer by positively regulating DNA binding and stability of AR-V7
Oncogene (2018)
-
A novel crosstalk between CCAR2 and AKT pathway in the regulation of cancer cell proliferation
Cell Death & Disease (2016)
-
Positive regulation of β-catenin–PROX1 signaling axis by DBC1 in colon cancer progression
Oncogene (2016)
-
DBC1/CCAR2 is involved in the stabilization of androgen receptor and the progression of osteosarcoma
Scientific Reports (2015)