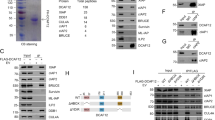
Abstract
The Drosophila melanogaster inhibitor of apoptosis protein DIAP1 suppresses apoptosis in part through inhibition of the effector caspase DrICE. The pro-death proteins Reaper, Hid and Grim (RHG) induce apoptosis by antagonizing DIAP1 function. However, the underlying molecular mechanisms remain unknown. Here we demonstrate that DIAP1 directly inhibits the catalytic activity of DrICE through its BIR1 domain and this inhibition is countered effectively by the RHG proteins. Inhibition of DrICE by DIAP1 occurs only after the cleavage of its N-terminal 20 amino acids and involves a conserved surface groove on BIR1. Crystal structures of BIR1 bound to the RHG peptides show that the RHG proteins use their N-terminal IAP-binding motifs to bind to the same surface groove, hence relieving DIAP1-mediated inhibition of DrICE. These studies define novel molecular mechanisms for the inhibition and activation of a representative D. melanogaster effector caspase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Shi, Y. Mechanisms of caspase inhibition and activation during apoptosis. Mol. Cell 9, 459– 470 (2002).
Thornberry, N.A. & Lazebnik, Y. Caspases: Enemies within. Science 281, 1312– 1316 (1998).
Deveraux, Q.L. & Reed, J.C. IAP family proteins—suppressors of apoptosis. Genes Dev. 13, 239– 252 (1999).
Salvesen, G.S. & Duckett, C.S. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3, 401– 410 (2002).
Hay, B.A., Wassarman, D.A. & Rubin, G.M. D. melanogaster homologs of baculoviral inhibitor of apoptosis proteins function to block cell death. Cell 83, 1253– 1262 (1995).
Shi, Y. A conserved tetrapeptide motif: Potentiating apoptosis through IAP-binding. Cell Death Differ. 9, 93– 95 (2002).
Shiozaki, E.N. et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519– 527 (2003).
Srinivasula, S.M. et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO mediates opposing effects on caspase activity and apoptosis. Nature 409, 112– 116 (2001).
Chai, J. et al. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406, 855– 862 (2000).
Wu, G. et al. Structural basis of IAP recognition by Smac/DIABLO. Nature 408, 1008– 1012 (2000).
Liu, Z. et al. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 408, 1004– 1008 (2000).
Chai, J. et al. Structural basis of caspase-7 inhibition by XIAP. Cell 104, 769– 780 (2001).
Huang, Y. et al. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell 104, 781– 790 (2001).
Riedl, S.J. et al. Structural basis for the inhibition of caspase-3 by XIAP. Cell 104, 791– 800 (2001).
Chai, J. et al. Molecular mechanism of Reaper/Grim/Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 10, 892– 898 (2003).
Wilson, R. et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4, 445– 450 (2002).
Fraser, A.G. & Evan, G.I. Identification of a D. melanogaster ICE/CED-3-related protease, drICE. EMBO J. 16, 2805– 2813 (1997).
Fraser, A.G., McCarthy, N.J. & Evan, G.I. drICE is an essential caspase required for apoptotic activity in D. melanogaster cells. EMBO J. 16, 6192– 6199 (1997).
Hawkins, C.J. et al. The D. melanogaster caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 275, 27084– 27093 (2000).
Meier, P., Silke, J., Leevers, S.J. & Evan, G.I. The D. melanogaster caspase DRONC is regulated by DIAP1. EMBO J. 19, 598– 611 (2000).
Kaiser, W., Vucic, D. & Miller, L. The D. melanogaster inhibitor of apoptosis DIAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 440, 243– 248 (1998).
Goyal, L., McCall, K., Agapite, J., Hartwieg, E. & Steller, H. Induction of apoptosis by D. melanogaster reaper, hid and grim through inhibition of IAP function. EMBO J. 19, 589– 597 (2000).
Wang, S., Hawkins, C., Yoo, S., Muller, H.-A. & Hay, B. The D. melanogaster caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98, 453– 463 (1999).
Lisi, S., Mazzon, I. & White, K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in D. melanogaster . Genetics 154, 669– 678 (2000).
Vucic, D., Kaiser, W.J. & Miller, L.K. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by D. melanogaster proteins HID and GRIM. Mol. Cell. Biol. 18, 3300– 3309 (1998).
Song, Z. et al. Biochemical and genetic interactions between D. melanogaster caspases and the proapoptotic genes rpr, hid, and grim. Mol. Cell Biol. 20, 2907– 2914 (2000).
Hawkins, C., Wang, S. & Hay, B.A. A cloning method to identify caspases and their regulators in yeast: identification of D. melanogaster IAP1 as an inhibitor of the D. melanogaster caspase DCP-1. Proc. Natl. Acad. Sci. USA 96, 2885– 2890 (1999).
Vucic, D., Kaiser, W.J., Harvey, A.J. & Miller, L.K. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA 94, 10183– 10188 (1997).
Dorstyn, L., Colussi, P.A., Quinn, L.M., Richardson, H. & Kumar, S. DRONC, an ecdysone-inducible D. melanogaster caspase. Proc. Natl. Acad. Sci. USA 96, 4307– 4312 (1999).
Sun, C. et al. NMR structure and mutagenesis of the third BIR domain of the inhibitor of apoptosis protein XIAP. J. Biol. Chem. 275, 33777– 33781 (2000).
Fesik, S.W. Insights into programmed cell death through structural biology. Cell 103, 273– 282 (2000).
Wu, J.-W., Cocina, A.E., Chai, J., Hay, B.A. & Shi, Y. Structural analysis of a functional DIAP1 fragment bound to Grim and Hid peptides. Mol. Cell 8, 95– 104 (2001).
Vernooy, S.Y. et al. Cell death regulation in D. melanogaster: conservation of mechanism and unique insights. J. Cell Biol. 150, F69– F76 (2000).
Ditzel, M. et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5, 467– 473 (2003).
Zachariou, A. et al. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 22, 6642– 6652 (2003).
Chai, J. et al. Crystal Structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell 107, 399– 407 (2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307– 326 (1997).
Navaza, J. AMoRe and automated package for molecular replacement. Acta Crystallogr. A 50, 157– 163 (1994).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110– 119 (1991).
Terwilliger, T.C. & Berendzen, J. Correlated phasing of multiple isomorphous replacement data. Acta Crystallogr. D 52, 749– 757 (1996).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946– 950 (1991).
Acknowledgements
We thank L. Walsh at CHESS for help with beam time and T.K. Vanderlick and M. Apel-Paz for help with ITC. This work was supported by US National Institutes of Health grants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Yan, N., Wu, JW., Chai, J. et al. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol 11, 420–428 (2004). https://doi.org/10.1038/nsmb764
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb764
This article is cited by
-
An inhibitor of apoptosis protein (EsIAP1) from Chinese mitten crab Eriocheir sinensis regulates apoptosis through inhibiting the activity of EsCaspase-3/7-1
Scientific Reports (2019)
-
Loss of putzig in the germline impedes germ cell development by inducing cell death and new niche like microenvironments
Scientific Reports (2019)
-
Engineered XylE as a tool for mechanistic investigation and ligand discovery of the glucose transporters GLUTs
Cell Discovery (2019)
-
Cryo-EM structure of Nma111p, a unique HtrA protease composed of two protease domains and four PDZ domains
Cell Research (2017)
-
Genetic characterization of two gain-of-function alleles of the effector caspase DrICE in Drosophila
Cell Death & Differentiation (2016)