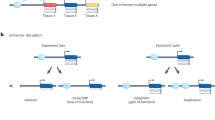
Abstract
Enhancers are cis-regulatory elements that enable precise spatiotemporal patterns of gene expression during development and are notable for being able to function at large distances from their target genes. Such regulatory elements often bypass intervening genes and typically comprise binding sites for multiple transcription factors that can also be transcribed by RNA polymerase II (Pol II) to produce noncoding enhancer RNAs (eRNAs). Genome-wide analyses have revealed chromatin signatures of enhancers, such as the enrichment for monomethylation of histone H3 lysine 4 (H3K4me1) and the acetylation or methylation of histone H3 lysine 27 (H3K27). Enhancer signatures have been used to describe the transitions of these regulatory elements from inactive to primed and from activated to decommissioned states during development. New mutations of enhancer sequences and of the protein factors regulating enhancer function in human disease continue to be identified, contributing to a growing class of 'enhanceropathies'.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Katie Vicari
Similar content being viewed by others
References
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981).
Moreau, P. et al. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 9, 6047–6068 (1981). Refs. 1 and 2 reported the identification of the SV40 enhancer whose properties became defining features of enhancers for the next 30 years.
Banerji, J., Olson, L. & Schaffner, W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729–740 (1983).
Gillies, S.D., Morrison, S.L., Oi, V.T. & Tonegawa, S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33, 717–728 (1983).
Mercola, M., Wang, X.F., Olsen, J. & Calame, K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science 221, 663–665 (1983). Refs. 3–5 reported the first cellular enhancer, which, when placed in proximity to the MYC gene, leads to Burkitt's lymphoma.
Taub, R. et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 79, 7837–7841 (1982).
Schroeder, M.D., Greer, C. & Gaul, U. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development 138, 3067–3078 (2011).
Levine, M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010).
Klingler, M., Soong, J., Butler, B. & Gergen, J.P. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev. Biol. 177, 73–84 (1996).
Jack, J., Dorsett, D., Delotto, Y. & Liu, S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113, 735–747 (1991).
Spana, C., Harrison, D.A. & Corces, V.G. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2, 1414–1423 (1988). This study showed that Su(Hw) directly bound sequences in the gypsy insulator, thus providing a mechanistic understanding for years of genetic studies and helping to launch the new field of insulator biology.
Morcillo, P., Rosen, C., Baylies, M.K. & Dorsett, D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11, 2729–2740 (1997).
Morcillo, P., Rosen, C. & Dorsett, D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics 144, 1143–1154 (1996). This study used a genetic screen for factors regulating enhancer-promoter communication and identified genes later shown to encode the fly homologs of NIPBL and LDB1. These proteins were subsequently shown to be involved in the formation and/or maintenance of enhancer-promoter looping in mammals.
Rollins, R.A., Korom, M., Aulner, N., Martens, A. & Dorsett, D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24, 3100–3111 (2004).
Rollins, R.A., Morcillo, P. & Dorsett, D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152, 577–593 (1999).
Agulnick, A.D. et al. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384, 270–272 (1996).
Lee, S.K. & Pfaff, S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731–745 (2003).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012). This study provided direct experimental confirmation that LDB1 bridges enhancer-promoter communication.
Torigoi, E. et al. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl. Acad. Sci. USA 97, 2686–2691 (2000).
Soler, E. et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 24, 277–289 (2010).
Kagey, M.H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). This study used genome-wide analyses and chromatin conformation assays to reveal the general requirement of cohesin in enhancer-promoter communication in mouse embryonic stem cells.
Schaaf, C.A. et al. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 9, e1003382 (2013).
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Kulaeva, O.I., Nizovtseva, E.V., Polikanov, Y.S., Ulianov, S.V. & Studitsky, V.M. Distant activation of transcription: mechanisms of enhancer action. Mol. Cell. Biol. 32, 4892–4897 (2012).
Williams, T.N. & Weatherall, D.J. World distribution, population genetics, and health burden of the hemoglobinopathies. Cold Spring Harb. Perspect. Med. 2, a011692 (2012).
Kioussis, D., Vanin, E., deLange, T., Flavell, R.A. & Grosveld, F.G. β-globin gene inactivation by DNA translocation in γβ-thalassaemia. Nature 306, 662–666 (1983).
Van der Ploeg, L.H. et al. γβ-Thalassaemia studies showing that deletion of the γ- and δ-genes influences β-globin gene expression in man. Nature 283, 637–642 (1980). Refs. 26 and 27 helped link a deletion in the DNase-hypersensitive β-globin locus control region with thalassemias.
Kioussis, D. & Festenstein, R. Locus control regions: overcoming heterochromatin-induced gene inactivation in mammals. Curr. Opin. Genet. Dev. 7, 614–619 (1997).
Fraser, P. & Grosveld, F. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 10, 361–365 (1998).
Gilbert, S.F. Developmental Biology (Sinauer Associates, Sunderland, Massachusetts, USA, 2000).
Bonifer, C., Vidal, M., Grosveld, F. & Sippel, A.E. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 9, 2843–2848 (1990).
Jones, B.K., Monks, B.R., Liebhaber, S.A. & Cooke, N.E. The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15, 7010–7021 (1995).
modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Kvon, E.Z., Stampfel, G., Yanez-Cuna, J.O., Dickson, B.J. & Stark, A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 26, 908–913 (2012).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Whyte, W.A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Koch, F. et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18, 956–963 (2011).
Parker, S.C. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA 110, 17921–17926 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Barolo, S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays 34, 135–141 (2012).
Lagha, M., Bothma, J.P. & Levine, M. Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 (2012).
Perry, M.W., Boettiger, A.N., Bothma, J.P. & Levine, M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562–1567 (2010).
Hong, J.W., Hendrix, D.A. & Levine, M.S. Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 (2008). This study used genome-wide occupancy of transcription factors in Drosophila to identify novel, seemingly redundant enhancers for well-known enhancers of developmental genes.
Frankel, N. et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010).
Casellas, R., Yamane, A., Kovalchuk, A.L. & Potter, M. Restricting activation-induced cytidine deaminase tumorigenic activity in B lymphocytes. Immunology 126, 316–328 (2009).
Kieffer-Kwon, K.R. et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155, 1507–1520 (2013). This study found that deletion of either the E1 or E2 enhancers for the Aicd gene led to the abolishment of Aicd expression. The authors suggested that genes whose induction needs to be tightly controlled require two enhancers to be turned on, a system referred to in this Review as fail-safe or split enhancers.
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Creyghton, M.P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). Refs. 47–49 used genome-wide mapping of histone modifications to identify chromatin signatures for different states of enhancer activity.
Whyte, W.A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012).
Herz, H.M. et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26, 2604–2620 (2012).
Hu, D. et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 (2013). Refs. 51 and 52 identified Drosophila Trr and its mammalian homologs MLL3 and MLL4 as major H3K4 monomethylases at enhancers.
Shilatifard, A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012).
Ardehali, M.B. et al. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30, 2817–2828 (2011).
Mohan, M. et al. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31, 4310–4318 (2011).
Eissenberg, J.C. & Shilatifard, A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249 (2010).
Sedkov, Y. et al. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech. Dev. 82, 171–179 (1999).
Wang, P. et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29, 6074–6085 (2009).
Yu, B.D., Hess, J.L., Horning, S.E., Brown, G.A. & Korsmeyer, S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995).
Tie, F. et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 (2009).
Jin, Q. et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011).
Wang, L., Tang, Y., Cole, P.A. & Marmorstein, R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747 (2008).
Roelfsema, J.H. et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 76, 572–580 (2005).
Roth, J.F. et al. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 22, 5186–5196 (2003).
Kharchenko, P.V. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011).
Di Croce, L. & Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013).
Schwartz, Y.B. & Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22 (2007).
Herz, H.M., Garruss, A. & Shilatifard, A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639 (2013).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Tie, F., Banerjee, R., Conrad, P.A., Scacheri, P.C. & Harte, P.J. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol. Cell. Biol. 32, 2323–2334 (2012).
Lee, J.S., Smith, E. & Shilatifard, A. The language of histone crosstalk. Cell 142, 682–685 (2010).
Thornton, J.L. et al. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 28, 115–120 (2014).
Natoli, G. & Andrau, J.C. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46, 1–19 (2012).
Ørom, U.A. & Shiekhattar, R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193 (2013).
Kim, T.K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
De Santa, F. et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384 (2010).
Ashe, H.L., Monks, J., Wijgerde, M., Fraser, P. & Proudfoot, N.J. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 11, 2494–2509 (1997).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Ørom, U.A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010). Refs. 78 and 79 showed that some of the most active enhancers are transcribed.
Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013).
Wang, K.C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). Refs. 79–81 found that polyadenylated lncRNAs could activate promoters of nearby genes. When placed in reporter constructs, these RNAs have enhancer-like function, revealing a previously unknown diversity in the cis -regulatory landscape.
Rinn, J.L. & Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Zaret, K.S. & Carroll, J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Liber, D. et al. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell 7, 114–126 (2010).
Xu, J. et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838 (2009).
Lin, C., Garruss, A.S., Luo, Z., Guo, F. & Shilatifard, A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152, 144–156 (2013).
Hou, C., Dale, R. & Dean, A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. USA 107, 3651–3656 (2010).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Kaikkonen, M.U. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013). This study showed that the ongoing process of transcription at an enhancer, but not the eRNA product, was required for enhancer function.
Liu, J. & Krantz, I.D. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 9, 303–320 (2008).
Dorsett, D. & Krantz, I.D. On the molecular etiology of Cornelia de Lange syndrome. Ann. NY Acad. Sci. 1151, 22–37 (2009).
Borck, G. et al. Father-to-daughter transmission of Cornelia de Lange syndrome caused by a mutation in the 5′ untranslated region of the NIPBL gene. Hum. Mutat. 27, 731–735 (2006).
Ng, S.B. et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 (2010).
Lederer, D. et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 90, 119–124 (2012).
Miyake, N. et al. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am. J. Med. Genet. A. 161, 2234–2243 (2013).
Herz, H.M. et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 30, 2485–2497 (2010).
Kanda, H., Nguyen, A., Chen, L., Okano, H. & Hariharan, I.K. The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as a negative regulator of tissue growth. Mol. Cell. Biol. 33, 1702–1710 (2013).
Morgan, M.A. & Shilatifard, A. Drosophila SETs its sights on cancer: Trr/MLL3/4 COMPASS-like complexes in development and disease. Mol. Cell. Biol. 33, 1698–1701 (2013).
Grasso, C.S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Jones, D.T. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Morin, R.D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Parsons, D.W. et al. The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Pugh, T.J. et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012).
Akhtar-Zaidi, B. et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336, 736–739 (2012).
Das, C., Lucia, M.S., Hansen, K.C. & Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Wang, F., Marshall, C.B. & Ikura, M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell. Mol. Life Sci. 70, 3989–4008 (2013).
Zheng, R. & Blobel, G.A. GATA transcription factors and cancer. Genes Cancer 1, 1178–1188 (2010).
Hsu, A.P. et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118, 2653–2655 (2011).
Hsu, A.P. et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121, 3830–3837 (2013).
Lettice, L.A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003).
Lettice, L.A., Hill, A.E., Devenney, P.S. & Hill, R.E. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum. Mol. Genet. 17, 978–985 (2008).
Corradin, O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24, 1–13 (2013).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Encode Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Bauer, D.E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013).
Xu, J. et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 334, 993–996 (2011). These authors deleted a tissue-specific enhancer of an essential gene encoding a transcriptional repressor for fetal hemoglobin and found that this disruption restored globin expression in a mouse model of sickle-cell anemia.
Pastor, W.A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Arnold, C.D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013). This study developed a genome-wide screen to identify regions of DNA that function in enhancer assays in Drosophila S2 cells.
Gaj, T., Gersbach, C.A. & Barbas, C.F. III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013).
Mendenhall, E.M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31, 1133–1136 (2013). This study used TAL effectors to target LSD1 to silence an enhancer. This approach can help determine the in vivo function of an enhancer and could potentially be used for therapeutic purposes.
Li, L.M. & Arnosti, D.N. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr. Biol. 21, 406–412 (2011).
Li, L., Greer, C., Eisenman, R.N. & Secombe, J. Essential functions of the histone demethylase lid. PLoS Genet. 6, e1001221 (2010).
Morales Torres, C., Laugesen, A. & Helin, K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS ONE 8, e60020 (2013).
Shpargel, K.B., Sengoku, T., Yokoyama, S. & Magnuson, T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 (2012).
Terranova, R., Agherbi, H., Boned, A., Meresse, S. & Djabali, M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA 103, 6629–6634 (2006).
Wang, C. et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. USA 109, 15324–15329 (2012).
Lam, M.T.Y. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 (2013). This study demonstrated that an eRNA product has regulatory function.
Le Borgne, R., Remaud, S., Hamel, S. & Schweisguth, F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3, e96 (2005).
Chlon, T.M. & Crispino, J.D. Combinatorial regulation of tissue specification by GATA and FOG factors. Development 139, 3905–3916 (2012).
Cirillo, L.A. et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 17, 244–254 (1998). This study demonstrated that the pioneer factor HNF3 not only has a structure resembling that of linker histone H1, but binds to nucleosomal DNA in a similar manner.
Cuesta, I., Zaret, K.S. & Santisteban, P. The forkhead factor FoxE1 binds to the thyroperoxidase promoter during thyroid cell differentiation and modifies compacted chromatin structure. Mol. Cell. Biol. 27, 7302–7314 (2007).
Hatta, M. & Cirillo, L.A. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J. Biol. Chem. 282, 35583–35593 (2007).
Mandel, E.M. & Grosschedl, R. Transcription control of early B cell differentiation. Curr. Opin. Immunol. 22, 161–167 (2010).
Natoli, G., Ghisletti, S. & Barozzi, I. The genomic landscapes of inflammation. Genes Dev. 25, 101–106 (2011).
Harrison, M.M., Li, X.Y., Kaplan, T., Botchan, M.R. & Eisen, M.B. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266 (2011).
Liang, H.L. et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008).
Nien, C.Y. et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339 (2011).
Dorsett, D. & Merkenschlager, M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol. 25, 327–333 (2013).
Matthews, J.M. & Visvader, J.E. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 4, 1132–1137 (2003).
Ghirlando, R. et al. Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta 1819, 644–651 (2012).
Chetverina, D., Aoki, T., Erokhin, M., Georgiev, P. & Schedl, P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays 36, 163–172 (2014).
Phillips-Cremins, J.E. & Corces, V.G. Chromatin insulators: linking genome organization to cellular function. Mol. Cell 50, 461–474 (2013).
Guo, C. et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget 4, 2144–2153 (2013).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Krebs, A.R., Karmodiya, K., Lindahl-Allen, M., Struhl, K. & Tora, L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell 44, 410–423 (2011).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009).
Shi, J. et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 27, 2648–2662 (2013).
Belkina, A.C. & Denis, G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 12, 465–477 (2012).
Acknowledgements
We thank D. Dorsett and H.-M. Herz for helpful discussions, M. Miller for preparation of figures, M. Morgan for the term 'enhanceropathy', and L. Shilatifard and L. Kennedy for editorial assistance. The work on enhancers in the Shilatifard laboratory is funded by US National Institutes of Health grants R01CA150265 and R01CA89455.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Smith, E., Shilatifard, A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21, 210–219 (2014). https://doi.org/10.1038/nsmb.2784
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2784
This article is cited by
-
A multiple super-enhancer region establishes inter-TAD interactions and controls Hoxa function in cranial neural crest
Nature Communications (2023)
-
G-quadruplexes sense natural porphyrin metabolites for regulation of gene transcription and chromatin landscapes
Genome Biology (2022)
-
StackEPI: identification of cell line-specific enhancer–promoter interactions based on stacking ensemble learning
BMC Bioinformatics (2022)
-
Comprehensive enhancer-target gene assignments improve gene set level interpretation of genome-wide regulatory data
Genome Biology (2022)
-
A map of cis-regulatory modules and constituent transcription factor binding sites in 80% of the mouse genome
BMC Genomics (2022)