Abstract
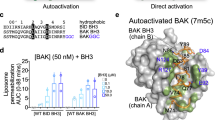
The BCL-2–family protein BAK is responsible for mitochondrial outer-membrane permeabilization (MOMP), which leads to apoptosis. The BCL-2 homology 3 (BH3)-only protein BID activates BAK to perform this function. We report the NMR solution structure of the human BID BH3–BAK complex, which identified the activation site at the canonical BH3-binding groove of BAK. Mutating the BAK BH1 in the groove prevented activation and MOMP but not the binding of BID. BAK BH3 mutations allowed BID binding and activation but blunted function by blocking BAK oligomerization. BAK activation follows a 'hit-and-run' mechanism whereby BID dissociates from the trigger site, which allows BAK oligomerization at an overlapping interface. In contrast, the BH3-only proteins NOXA and BAD are predicted to clash with the trigger site and are not activators of BAK. These findings provide insights into the early stages of BAK activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Green, D.R. Apoptotic pathways: ten minutes to dead. Cell 121, 671–674 (2005).
Jiang, X. & Wang, X. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 (2004).
Tait, S.W. & Green, D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 (2010).
Kuwana, T. et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342 (2002).
Lindsten, T. et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell 6, 1389–1399 (2000).
Wei, M.C. et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 (2001).
Dewson, G. et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell 30, 369–380 (2008).
Dewson, G. et al. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 19, 661–670 (2012).
Chipuk, J.E., Moldoveanu, T., Llambi, F., Parsons, M.J. & Green, D.R. The BCL-2 family reunion. Mol. Cell 37, 299–310 (2010).
Llambi, F. et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44, 517–531 (2011).
Chipuk, J.E. & Green, D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 18, 157–164 (2008).
Walensky, L.D. & Gavathiotis, E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem. Sci. 36, 642–652 (2011).
Westphal, D., Dewson, G., Czabotar, P.E. & Kluck, R.M. Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta 1813, 521–531 (2011).
Certo, M. et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9, 351–365 (2006).
Bird, G.H., Bernal, F., Pitter, K. & Walensky, L.D. Synthesis and biophysical characterization of stabilized α-helices of BCL-2 domains. Methods Enzymol. 446, 369–386 (2008).
Gavathiotis, E., Reyna, D.E., Davis, M.L., Bird, G.H. & Walensky, L.D. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell 40, 481–492 (2010).
Gavathiotis, E. et al. BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 (2008).
Moldoveanu, T. et al. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol. Cell 24, 677–688 (2006).
Kim, Y.W., Grossmann, T.N. & Verdine, G.L. Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat. Protoc. 6, 761–771 (2011).
Walensky, L.D. et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004).
Gräslund, S. et al. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr. Purif. 58, 210–221 (2008).
Wang, H. et al. Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator. J. Struct. Biol. 166, 32–37 (2009).
Gross, A. et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 274, 1156–1163 (1999).
Ku, B., Liang, C., Jung, J.U. & Oh, B.H. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 21, 627–641 (2011).
Letai, A.G. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat. Rev. Cancer 8, 121–132 (2008).
Wei, M.C. et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14, 2060–2071 (2000).
Oltersdorf, T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005).
Dai, H. et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J. Cell Biol. 194, 39–48 (2011).
Strasser, A., Jost, P.J. & Nagata, S. The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192 (2009).
Kim, H. et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 36, 487–499 (2009).
Ren, D. et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 330, 1390–1393 (2010).
Du, H. et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J. Biol. Chem. 286, 491–501 (2011).
Czabotar, P.E. et al. Bax crystal structures reveal how BH3 domains activate bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 (2013).
Leshchiner, E.S., Braun, C.R., Bird, G.H. & Walensky, L.D. Direct activation of full-length proapoptotic BAK. Proc. Natl. Acad. Sci. USA 110, E986–E995 (2013).
Ferrer, P.E., Frederick, P., Gulbis, J.M., Dewson, G. & Kluck, R.M. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome C release: implications for Bak and Bax apoptotic function. PLoS ONE 7, e31510 (2012).
Oh, K.J. et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3–BH3 contact interface in BAK homo-oligomers. J. Biol. Chem. 285, 28924–28937 (2010).
Dewson, G. et al. Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol. Cell 36, 696–703 (2009).
Lovell, J.F. et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135, 1074–1084 (2008).
Edlich, F. et al. Bcl-xL retrotranslocates Bax from the mitochondria into the cytosol. Cell 145, 104–116 (2011).
Montessuit, S. et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142, 889–901 (2010).
Liu, X. et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal. Biochem. 280, 20–28 (2000).
Denisov, A.Y. et al. Structural model of the BCL-w-BID peptide complex and its interactions with phospholipid micelles. Biochemistry 45, 2250–2256 (2006).
Moldoveanu, T., Gehring, K. & Green, D.R. Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature 456, 404–408 (2008).
Moldoveanu, T. et al. A Ca2+ switch aligns the active site of calpain. Cell 108, 649–660 (2002).
Verdine, G.L. & Hilinski, G.J. Stapled peptides for intracellular drug targets. Methods Enzymol. 503, 3–33 (2012).
Hill, J.M. NMR screening for rapid protein characterization in structural proteomics. Methods Mol. Biol. 426, 437–446 (2008).
Sattler, M., Schleucher, J. & Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 (1999).
Keller, R. The computer aided resonance assignment tutorial. (CANTINA Verlag, Goldau, Switzerland, 2004).
Guerry, P. & Herrmann, T. Comprehensive automation for NMR structure determination of proteins. Methods Mol. Biol. 831, 429–451 (2012).
Güntert, P., Mumenthaler, C. & Wuthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Laue, T.M., Shah, B.D., Ridgeway, T.M. & Pelletier, S.L. Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds. Harding, S.E., Rowe, A.J. & Horton, J.C.) 90–125 (The Royal Society of Chemistry, 1992).
Brown, P.H., Balbo, A. & Schuck, P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. 81, 18.15 (2008).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Balbo, A., Brown, P.H., Braswell, E.H. & Schuck, P. Measuring protein-protein interactions by equilibrium sedimentation. Curr. Protoc. Immunol. 81, 18.8 (2007).
Acknowledgements
We thank C. Dillon, L. McCormick and M. Yang for managing the mouse colony, R. Cassell and P. Rodriques of the St. Jude Hartwell Center for producing the peptides and R. Cross and G. Lennon of the St. Jude Flow Cytometry facility for cell sorting. This work was supported by US National Institutes of Health grants AI40646, GM52735 and GM096208 (D.R.G.) and R01CA082491 and 1R01GM083159 (R.W.K.) and by the American Lebanese Syrian Associated Charities.
Author information
Authors and Affiliations
Contributions
T.M., C.R.G., K.G. and R.W.K. contributed the NMR analyses, T.M., F.L. and P.F. the biochemical analyses and T.M. and A.N. the AUC analyses. All authors contributed conceptually to various aspects of the project. T.M., R.W.K. and D.R.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–2 and Supplementary Note (PDF 1699 kb)
Supplementary Movie 1
Movie captures the conformational changes of apo to SAHBa–bound cBAK described in Fig. 2 andSupplementary Fig. 2. LSQMAN3 was used to generate the frames and Pymol to render them. (AVI 9300 kb)
Supplementary Movie 2
Movie captures the conformational changes of apo to SAHBa–bound cBAK from a side viewcompared to that of Supplementary Movie 1. (AVI 12207 kb)
Rights and permissions
About this article
Cite this article
Moldoveanu, T., Grace, C., Llambi, F. et al. BID-induced structural changes in BAK promote apoptosis. Nat Struct Mol Biol 20, 589–597 (2013). https://doi.org/10.1038/nsmb.2563
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2563
This article is cited by
-
A novel inhibitory BAK antibody enables assessment of non-activated BAK in cancer cells
Cell Death & Differentiation (2024)
-
Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove
Communications Biology (2023)
-
Eltrombopag directly activates BAK and induces apoptosis
Cell Death & Disease (2023)
-
Targeting Cpt1a-Bcl-2 interaction modulates apoptosis resistance and fibrotic remodeling
Cell Death & Differentiation (2022)
-
Structural basis of BAK activation in mitochondrial apoptosis initiation
Nature Communications (2022)