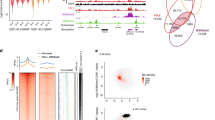
Abstract
Transcriptional activation is controlled by chromatin, which needs to be unfolded and remodeled to ensure access to the transcription start site (TSS). However, the mechanisms that yield such an 'open' chromatin structure, and how these processes are coordinately regulated during differentiation, are poorly understood. We identify the mouse (Mus musculus) H2A histone variant H2A.Lap1 as a previously undescribed component of the TSS of active genes expressed during specific stages of spermatogenesis. This unique chromatin landscape also includes a second histone variant, H2A.Z. In the later stages of round spermatid development, H2A.Lap1 dynamically loads onto the inactive X chromosome, enabling the transcriptional activation of previously repressed genes. Mechanistically, we show that H2A.Lap1 imparts unique unfolding properties to chromatin. We therefore propose that H2A.Lap1 coordinately regulates gene expression by directly opening the chromatin structure of the TSS at genes regulated during spermatogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
11 December 2011
In the version of this article initially published, the incorrect PDB code for the MthK open channel structure was provided in the legend to Figure 1. The correct PDB code for this structure is 1LNQ. The error has been corrected in the HTML and PDF versions of the article.
References
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Caterino, T.L. & Hayes, J.J. Chromatin structure depends on what's in the nucleosome's pocket. Nat. Struct. Mol. Biol. 14, 1056–1058 (2007).
Zhou, J., Fan, J.Y., Rangasamy, D. & Tremethick, D.J. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14, 1070–1076 (2007).
Ishibashi, T. et al. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 38, 1780–1789 (2009).
Greaves, I.K., Rangasamy, D., Devoy, M., Marshall Graves, J.A. & Tremethick, D.J. The X and Y chromosomes assemble into H2A.Z-containing facultative heterochromatin following meiosis. Mol. Cell. Biol. 26, 5394–5405 (2006).
Namekawa, S.H. et al. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16, 660–667 (2006).
Turner, J.M., Mahadevaiah, S.K., Ellis, P.J., Mitchell, M.J. & Burgoyne, P.S. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10, 521–529 (2006).
Turner, J.M., Burgoyne, P.S. & Singh, P.B. M31 and macroH2A1.2 colocalise at the pseudoautosomal region during mouse meiosis. J. Cell Sci. 114, 3367–3375 (2001).
Oakberg, E.F. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am. J. Anat. 99, 391–413 (1956).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Schones, D.E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Wang, Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 (2008).
Conerly, M.L. et al. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 20, 1383–1390 (2010).
Henikoff, S., Henikoff, J.G., Sakai, A., Loeb, G.B. & Ahmad, K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 19, 460–469 (2009).
Jin, C. et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 (2009).
Hansen, J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31, 361–392 (2002).
Montel, F. et al. The dynamics of individual nucleosomes controls the chromatin condensation pathway: direct atomic force microscopy visualization of variant chromatin. Biophys. J. 97, 544–553 (2009).
Allahverdi, A. et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 39, 1680–1691 (2011).
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Angelov, D. et al. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 23, 3815–3824 (2004).
Peters, A.H., Plug, A.W., van Vugt, M.J. & de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214 (1997).
Acknowledgements
We thank J. Fan (The Australian National University) for initial protein preparations of H2A.Lap1, J. Pehrson (University of Pennsylvania) for macroH2A antibodies, K. Luger (Colorado State University) for core histone recombinant proteins, S. McBryant and J. Hansen for helping M.N. to set up sedimentation velocity experiments, S. Grigoryev and R. Reeves for reading the manuscript, our in-house Biomolecular Research Service, headed by S. Palmer, for high-throughput DNA sequencing, and A. Prins and C. Gillespie for help in histological sample preparation and microscopy. This work was supported by Australian National Health and Medical Research Council project grants to T.A.S. and D.J.T., and to M.N. and D.J.T.
Author information
Authors and Affiliations
Contributions
T.A.S. helped design the experiments, cloned H2A.Lap, conducted all spermatogenesis experiments, prepared chromatin for ChIP-seq experiments and conducted the gene expression and ChIP experiments on individual X-chromosome genes. M.N. conducted the biochemical and biophysical experiments on the nucleosome arrays and prepared DNA ChIP libraries for high-throughput sequencing. R.W. developed and did data analysis of global mouse gene expression data. G.A.H. designed and executed the analysis of the Illumina short-read data. A.P. assisted with the analyses of Illumina short-read data. D.J.T. conceived the project, helped design the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Methods (PDF 2300 kb)
Rights and permissions
About this article
Cite this article
Soboleva, T., Nekrasov, M., Pahwa, A. et al. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol 19, 25–30 (2012). https://doi.org/10.1038/nsmb.2161
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2161
This article is cited by
-
Histone variant H2A.Z modulates nucleosome dynamics to promote DNA accessibility
Nature Communications (2023)
-
Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome
Communications Biology (2021)
-
Short H2A histone variants are expressed in cancer
Nature Communications (2021)
-
Identification of differentially expressed genes in mouse embryonic stem cell under hypoxia
Genes & Genomics (2021)
-
The roles of histone variants in fine-tuning chromatin organization and function
Nature Reviews Molecular Cell Biology (2020)