Abstract
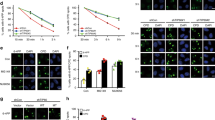
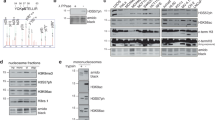
KAP-1 poses a substantial barrier to DNA double-strand break (DSB) repair within heterochromatin that is alleviated by ATM-dependent KAP-1 phosphorylation (pKAP-1). Here we address the mechanistic consequences of pKAP-1 that promote heterochromatic DSB repair and chromatin relaxation. KAP-1 function involves autoSUMOylation and recruitment of nucleosome deacetylation, methylation and remodeling activities. Although heterochromatin acetylation or methylation changes were not detected, radiation-induced pKAP-1 dispersed the nucleosome remodeler CHD3 from DSBs and triggered concomitant chromatin relaxation; pKAP-1 loss reversed these effects. Depletion or inactivation of CHD3, or ablation of its interaction with KAP-1SUMO1, bypassed pKAP-1's role in repair. Though KAP-1 SUMOylation was unaffected after irradiation, CHD3 dissociated from KAP-1SUMO1 in a pKAP-1–dependent manner. We demonstrate that KAP-1Ser824 phosphorylation generates a motif that directly perturbs interactions between CHD3′s SUMO-interacting motif and SUMO1, dispersing CHD3 from heterochromatin DSBs and enabling repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yunis, J.J. & Yasmineh, W.G. Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science 174, 1200–1209 (1971).
Miklos, G.L. & John, B. Heterochromatin and satellite DNA in man: properties and prospects. Am. J. Hum. Genet. 31, 264–280 (1979).
Craig, J.M. Heterochromatin—many flavours, common themes. Bioessays 27, 17–28 (2005).
Grewal, S.I. & Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007).
Becker, P.B. & Horz, W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273 (2002).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Hediger, F. & Gasser, S.M. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 16, 143–150 (2006).
Denslow, S.A. & Wade, P.A. The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 (2007).
Guenatri, M., Bailly, D., Maison, C. & Almouzni, G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 (2004).
Downs, J.A., Nussenzweig, M.C. & Nussenzweig, A. Chromatin dynamics and the preservation of genetic information. Nature 447, 951–958 (2007).
Falk, M., Lukasova, E. & Kozubek, S. Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat. Res. 704, 88–100 (2010).
Kim, J.A., Kruhlak, M., Dotiwala, F., Nussenzweig, A. & Haber, J.E. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 (2007).
Cowell, I.G. et al. γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE 2, e1057 (2007).
Goodarzi, A.A. et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 (2008).
Goodarzi, A.A., Jeggo, P. & Lobrich, M. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair (Amst.) 9, 1273–1282 (2010).
Ayoub, N., Jeyasekharan, A.D. & Venkitaraman, A.R. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle 8, 2945–2950 (2009).
Dinant, C. & Luijsterburg, M.S. The emerging role of HP1 in the DNA damage response. Mol. Cell. Biol. 29, 6335–6340 (2009).
Ziv, Y. et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 (2006).
Noon, A.T. et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12, 177–184 (2010).
Tjeertes, J.V., Miller, K.M. & Jackson, S.P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009).
Lee, Y.K., Thomas, S.N., Yang, A.J. & Ann, D.K. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J. Biol. Chem. 282, 1595–1606 (2007).
Li, X. et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177–36189 (2007).
Ivanov, A.V. et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 (2007).
Löbrich, M. et al. γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669 (2010).
Schultz, D.C., Friedman, J.R. & Rauscher, F.J. III . Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443 (2001).
Polo, S.E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010).
Smeenk, G. et al. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190, 741–749 (2010).
Larsen, D.H. et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 190, 731–740 (2010).
Riballo, E. et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16, 715–724 (2004).
Kerscher, O. SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 (2007).
Hecker, C.M., Rabiller, M., Haglund, K., Bayer, P. & Dikic, I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 (2006).
Stehmeier, P. & Muller, S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33, 400–409 (2009).
Miller, K.M. et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151 (2010).
Saether, T. et al. The chromatin remodeling factor Mi-2α acts as a novel co-activator for human c-Myb. J. Biol. Chem. 282, 13994–14005 (2007).
Seeler, J.S. et al. Common properties of nuclear body protein SP100 and TIF1α chromatin factor: role of SUMO modification. Mol. Cell. Biol. 21, 3314–3324 (2001).
Goodarzi, A.A. et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25, 3880–3889 (2006).
Acknowledgements
We thank O.S. Gabrielsen and T. Sæther (University of Oslo) for the CHD3 construct, A. Oliver (University of Sussex) for helpful structural biology discussions and Y. Ziv (University of Tel Aviv) for advice on MNase assays. A.A.G. was supported by a grant from the Association for International Cancer Research (AICR). The P.A.J. lab is funded by the Medical Research Council (UK), AICR, the Wellcome Research Trust and the Department of Health Radiation Protection Programme (UK).
Author information
Authors and Affiliations
Contributions
A.A.G. and T. K. conducted the experiments. A.A.G. and P.A.J. coauthored the manuscript and conceived of and designed the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Discussion (PDF 2272 kb)
Rights and permissions
About this article
Cite this article
Goodarzi, A., Kurka, T. & Jeggo, P. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18, 831–839 (2011). https://doi.org/10.1038/nsmb.2077
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2077
This article is cited by
-
HJURP is recruited to double-strand break sites and facilitates DNA repair by promoting chromatin reorganization
Oncogene (2024)
-
Mouse Chd4-NURD is required for neonatal spermatogonia survival and normal gonad development
Epigenetics & Chromatin (2022)
-
Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes
Cellular and Molecular Life Sciences (2021)
-
Targeting DNA repair in cancer: current state and novel approaches
Cellular and Molecular Life Sciences (2020)
-
The CHD6 chromatin remodeler is an oxidative DNA damage response factor
Nature Communications (2019)