Key Points
-
This paper describes the trafficking and functions of influenza A virus (IAV) viral ribonucleoproteins (vRNPs), which contain the genetic material of IAV, within the host cell. We emphasize how vRNPs interact with, and depend on, host factors and pathways, how vRNP structure contributes to its function and the key open questions that still need to be answered.
-
The structure of vRNPs in their native form is described.
-
The mechanism of vRNP nuclear import is explained, including a discussion of how vRNP components interact with the cellular importin-α–importin-β1 nuclear import pathway.
-
Primary genome transcription and genome replication is described, focusing on how host factors contribute to these processes.
-
Nuclear export of vRNP, which is mediated by the cellular CRM1 (also known as exportin-1) nuclear export pathway, is illustrated.
-
Recent work implicating the cellular RAB11 vesicle transport system and microtubules in vRNP cytoplasmic transport in late-stage infection is described.
-
We highlight important open questions and suggest methods that could be used to address these gaps in our knowledge.
Abstract
Influenza A viral ribonucleoprotein (vRNP) complexes comprise the eight genomic negative-sense RNAs, each of which is bound to multiple copies of the vRNP and a trimeric viral polymerase complex. The influenza virus life cycle centres on the vRNPs, which in turn rely on host cellular processes to carry out functions that are necessary for the successful completion of the virus life cycle. In this Review, we discuss our current knowledge about vRNP trafficking within host cells and the function of these complexes in the context of the virus life cycle, highlighting how structure contributes to function and the crucial interactions with host cell pathways, as well as on the information gaps that remain. An improved understanding of how vRNPs use host cell pathways is essential to identify mechanisms of virus pathogenicity, host adaptation and, ultimately, new targets for antiviral intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
World Health Organization. Influenza (Seasonal) Fact sheet N°211. WHO [online], (2009).
Molinari, N. A. et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 (2007).
Simonsen, L. The global impact of influenza on morbidity and mortality. Vaccine 17 (Suppl. 1), 3–10 (1999).
van der Vries, E., Schutten, M., Fraaij, P., Boucher, C. & Osterhaus, A. Influenza virus resistance to antiviral therapy. Adv. Pharmacol. 67, 217–246 (2013).
Matsuoka, Y. et al. A comprehensive map of the influenza A virus replication cycle. BMC syst. biol. 7, 97 (2013).
Shaw, M. L. & Palese, P. in Fields Virology. Section II: Specific Virus Families (eds Knipe, D. M. & Howley, P.) (Lippincott Williams and Wilkins, 2013).
Wright, P. F., Neumann, G. & Kawaoka, Y. in Fields Virology. Section II: Specific Virus Families (eds Knipe, D. M. & Howley, P.) (Lippincott Williams and Wilkins, 2013).
Gabriel, G. & Fodor, E. Molecular determinants of pathogenicity in the polymerase complex. Curr. Top. Microbiol. Immunol. 385, 35–60 (2014).
Scholtissek, C. & Becht, H. Binding of ribonucleic acids to the RNP-antigen protein of influenza viruses. J. General Virol. 10, 11–16 (1971).
Baudin, F., Bach, C., Cusack, S. & Ruigrok, R. W. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 13, 3158–3165 (1994).
Compans, R. W., Content, J. & Duesberg, P. H. Structure of the ribonucleoprotein of influenza virus. J. Virol. 10, 795–800 (1972).
Yamanaka, K., Ishihama, A. & Nagata, K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem. 265, 11151–11155 (1990).
Hsu, M. T., Parvin, J. D., Gupta, S., Krystal, M. & Palese, P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA 84, 8140–8144 (1987).
Fodor, E., Seong, B. L. & Brownlee, G. G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. General Virol. 74, 1327–1333 (1993).
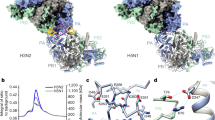
Arranz, R. et al. The structure of native influenza virion ribonucleoproteins. Science 338, 1634–1637 (2012). This paper describes the three-dimensional structure of native RNPs derived from influenza virions.
Moeller, A., Kirchdoerfer, R. N., Potter, C. S., Carragher, B. & Wilson, I. A. Organization of the influenza virus replication machinery. Science 338, 1631–1634 (2012). This paper describes the three-dimensional structure of native RNPs derived from cells expressing influenza RNP complex components.
Martin, K. & Helenius, A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67, 117–130 (1991). This paper was the first to establish that the influenza M1 protein regulates both nuclear import and nuclear export of influenza vRNPs.
Bui, M., Whittaker, G. & Helenius, A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70, 8391–8401 (1996).
Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nature Rev. Mol. Cell Biol. 8, 195–208 (2007).
Kutay, U., Bischoff, F. R., Kostka, S., Kraft, R. & Gorlich, D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071 (1997).
Martin, K. & Helenius, A. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65, 232–244 (1991). This paper was the first to show that influenza vRNPs enter the nucleus through the nuclear pore complex by an active mechanism.
Kemler, I., Whittaker, G. & Helenius, A. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology 202, 1028–1033 (1994).
O'Neill, R. E., Jaskunas, R., Blobel, G., Palese, P. & Moroianu, J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem. 270, 22701–22704 (1995).
Chou, Y. Y. et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 9, e1003358 (2013).
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Struct. Mol. Biol. 14, 229–233 (2007).
Nath, S. T. & Nayak, D. P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol. Cell. Biol. 10, 4139–4145 (1990).
Fodor, E. & Smith, M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78, 9144–9153 (2004).
Neumann, G., Castrucci, M. R. & Kawaoka, Y. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71, 9690–9700 (1997).
Wang, P., Palese, P. & O'Neill, R. E. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71, 1850–1856 (1997).
Weber, F., Kochs, G., Gruber, S. & Haller, O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250, 9–18 (1998).
Nieto, A., de la Luna, S., Barcena, J., Portela, A. & Ortin, J. Complex structure of the nuclear translocation signal of influenza virus polymerase PA subunit. J. General Virol. 75, 29–36 (1994).
Cros, J. F., Garcia-Sastre, A. & Palese, P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6, 205–213 (2005).
Gabriel, G. et al. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nature Commun. 2, 156 (2011).
Wu, W. W., Weaver, L. L. & Pante, N. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. J. Mol. Biol. 374, 910–916 (2007).
Gabriel, G., Herwig, A. & Klenk, H. D. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog. 4, e11 (2008).
Mark, G. E., Taylor, J. M., Broni, B. & Krug, R. M. Nuclear accumulation of influenza viral RNA transcripts and the effects of cycloheximide, actinomycin D, and α-amanitin. J. Virol. 29, 744–752 (1979).
Bouloy, M., Plotch, S. J. & Krug, R. M. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl Acad. Sci. USA 75, 4886–4890 (1978).
Plotch, S. J., Bouloy, M. & Krug, R. M. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl Acad. Sci. USA 76, 1618–1622 (1979).
Plotch, S. J., Bouloy, M., Ulmanen, I. & Krug, R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981).
Jorba, N., Coloma, R. & Ortin, J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5, e1000462 (2009). This paper describes a new model for influenza virus RNA transcription and replication. Specifically, the data suggest that primary transcription is carried out by the resident polymerase complex bound to the double-stranded end of the vRNP, whereas genome replication is carried out by soluble polymerase complexes and another polymerase complex directs the encapsidation of progeny vRNPs.
Robertson, J. S., Schubert, M. & Lazzarini, R. A. Polyadenylation sites for influenza virus mRNA. J. Virol. 38, 157–163 (1981).
Poon, L. L., Pritlove, D. C., Fodor, E. & Brownlee, G. G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73, 3473–3476 (1999).
Fodor, E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol. 57, 113–122 (2013).
Yanguez, E. & Nieto, A. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res. 156, 1–12 (2011).
Schneider, J. & Wolff, T. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine 27, 6312–6316 (2009).
Vreede, F. T., Jung, T. E. & Brownlee, G. G. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78, 9568–9572 (2004).
York, A., Hengrung, N., Vreede, F. T., Huiskonen, J. T. & Fodor, E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc. Natl Acad. Sci. USA 110, E4238–E4245 (2013).
Deng, T. et al. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80, 11911–11919 (2006).
Manzoor, R. et al. Heat shock protein 70 modulates Influenza A virus polymerase activity. J. Biol. Chem. http://dx.doi.org/10.1074/jbc.M113.507798 (2014).
Momose, F. et al. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277, 45306–45314 (2002).
Naito, T., Momose, F., Kawaguchi, A. & Nagata, K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81, 1339–1349 (2007).
Engelhardt, O. G., Smith, M. & Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79, 5812–5818 (2005).
Llompart, C. M., Nieto, A. & Rodriguez-Frandsen, A. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. J. Virol. 88, 3455–3463 (2014).
Landeras-Bueno, S., Jorba, N., Perez-Cidoncha, M. & Ortin, J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 7, e1002397 (2011).
Jorba, N. et al. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8, 2077–2088 (2008).
Kawaguchi, A. & Nagata, K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26, 4566–4575 (2007).
Naito, T. et al. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl Acad. Sci. USA 104, 18235–18240 (2007).
Momose, F. et al. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75, 1899–1908 (2001).
Kawaguchi, A., Momose, F. & Nagata, K. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. J. Virol. 85, 6197–6204 (2011).
Zhou, Z. et al. Fragile X mental retardation protein stimulates ribonucleoprotein assembly of influenza A virus. Nature Commun. 5, 3259 (2014).
Perez-Gonzalez, A., Rodriguez, A., Huarte, M., Salanueva, I. J. & Nieto, A. hCLE/CGI-99, a human protein that interacts with the influenza virus polymerase, is a mRNA transcription modulator. J. Mol. Biol. 362, 887–900 (2006).
Rodriguez, A., Perez-Gonzalez, A. & Nieto, A. Cellular human CLE/C14orf166 protein interacts with influenza virus polymerase and is required for viral replication. J. Virol. 85, 12062–12066 (2011).
Hudjetz, B. & Gabriel, G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-α1 and -α7. PLoS Pathog. 8, e1002488 (2012). This paper showed that IMPα1 and IMPα7 are positive regulators of human-like, but not avian-like, influenza polymerase activity, independent of IMP nuclear import activities, which suggests that IMPs regulate IAV transmission between reservoir species and humans.
Resa-Infante, P. et al. The host-dependent interaction of α-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE 3, e3904 (2008).
Shinya, K. et al. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320, 258–266 (2004).
Hatta, M., Gao, P., Halfmann, P. & Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293, 1840–1842 (2001).
Li, Z. et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79, 12058–12064 (2005).
Subbarao, E. K., London, W. & Murphy, B. R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67, 1761–1764 (1993).
Manz, B., Schwemmle, M. & Brunotte, L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 87, 7200–7209 (2013).
Shapira, S. D. et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 (2009).
Tafforeau, L. et al. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J. Virol. 85, 13010–13018 (2011).
Bradel-Tretheway, B. G. et al. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J. Virol. 85, 8569–8581 (2011).
Ma, K., Roy, A. M. & Whittaker, G. R. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282, 215–220 (2001).
Elton, D. et al. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75, 408–419 (2001).
Watanabe, K. et al. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77, 31–42 (2001).
Bui, M., Wills, E. G., Helenius, A. & Whittaker, G. R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74, 1781–1786 (2000).
Chase, G. P. et al. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 7, e1002187 (2011). This paper describes a strong association between influenza vRNPs and cellular chromatin, which precedes vRNP nuclear export in late-stage infection. The authors hypothesize that influenza virus uses this mechanism to gain preferential access to nuclear export machinery to promote vRNP nuclear export and, ultimately, replicative ability.
Kurokawa, M., Ochiai, H., Nakajima, K. & Niwayama, S. Inhibitory effect of protein kinase C inhibitor on the replication of influenza type A virus. J. General Virol. 71, 2149–2155 (1990).
Cao, S. et al. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J. Virol. 86, 4883–4891 (2012).
Sakaguchi, A., Hirayama, E., Hiraki, A., Ishida, Y. & Kim, J. Nuclear export of influenza viral ribonucleoprotein is temperature-dependently inhibited by dissociation of viral matrix protein. Virology 306, 244–253 (2003).
Zhirnov, O. P. & Klenk, H. D. Histones as a target for influenza virus matrix protein M1. Virology 235, 302–310 (1997).
Whittaker, G., Bui, M. & Helenius, A. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. J. Virol. 70, 2743–2756 (1996).
O'Neill, R. E., Talon, J. & Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17, 288–296 (1998). This paper was the first to establish that the influenza NEP protein regulates vRNP nuclear export and identify a region of the NEP protein that functions as a nuclear export signal.
Neumann, G., Hughes, M. T. & Kawaoka, Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19, 6751–6758 (2000). This paper established the importance of the NEP protein in the nuclear export of vRNPs in the context of viral infection and identified the most crucial residues of the NEP nuclear export signal.
Huang, S. et al. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 87, 767–778 (2013).
Akarsu, H. et al. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 22, 4646–4655 (2003).
Imai, M., Watanabe, S. & Odagiri, T. Influenza B virus NS2, a nuclear export protein, directly associates with the viral ribonucleoprotein complex. Arch. Virol. 148, 1873–1884 (2003).
Robb, N. C., Smith, M., Vreede, F. T. & Fodor, E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. General Virol. 90, 1398–1407 (2009).
Brunotte, L. et al. The nuclear export protein of H5N1 Influenza A viruses recruits matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J. Biol. Chem. 289, 20067–20077 (2014).
Shimizu, T., Takizawa, N., Watanabe, K., Nagata, K. & Kobayashi, N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS lett. 585, 41–46 (2011).
Yu, M. et al. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J. Virol. 86, 4970–4980 (2012).
Pleschka, S. et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nature Cell Biol. 3, 301–305 (2001). This paper established the importance of the RAF–MEK–ERK pathway to efficient influenza virus replication and identified the nuclear export of influenza vRNPs as the life cycle step that is affected by inhibition of RAF–MEK–ERK signalling, which indicates that cellular kinase pathways are required for vRNP nuclear export.
Kumar, N., Liang, Y. & Parslow, T. G. Receptor tyrosine kinase inhibitors block multiple steps of influenza A virus replication. J. Virol. 85, 2818–2827 (2011).
Alamares-Sapuay, J. G. et al. Serum- and glucocorticoid-regulated kinase 1 is required for nuclear export of the ribonucleoprotein of influenza A virus. J. Virol. 87, 6020–6026 (2013).
Marjuki, H. et al. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Cα-mediated activation of ERK signaling. J. Biol. Chem. 281, 16707–16715 (2006).
Marjuki, H. et al. Higher polymerase activity of a human influenza virus enhances activation of the hemagglutinin-induced Raf/MEK/ERK signal cascade. Virol. J. 4, 134 (2007).
Reinhardt, J. & Wolff, T. The influenza A virus M1 protein interacts with the cellular receptor of activated C kinase (RACK) 1 and can be phosphorylated by protein kinase C. Veterinary Microbiol. 74, 87–100 (2000).
Petri, T. & Dimmock, N. J. Phosphorylation of influenza virus nucleoprotein in vivo. J. Gen. Virol. 57, 185–190 (1981).
Kistner, O., Muller, K. & Scholtissek, C. Differential phosphorylation of the nucleoprotein of influenza A viruses. J. Gen. Virol. 70, 2421–2431 (1989).
Richardson, J. C. & Akkina, R. K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch. Virol. 116, 69–80 (1991).
Hutchinson, E. C. et al. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8, e1002993 (2012).
Takizawa, N., Kumakura, M., Takeuchi, K., Kobayashi, N. & Nagata, K. Sorting of influenza A virus RNA genome segments after nuclear export. Virology 401, 248–256 (2010).
Leser, G. P. & Lamb, R. A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342, 215–227 (2005).
Momose, F., Kikuchi, Y., Komase, K. & Morikawa, Y. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect. 9, 1422–1433 (2007).
Amorim, M. J. et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 85, 4143–4156 (2011). This paper was the first to identify overlap between the RAB11 recycling endosome compartment and the localization of cytoplasmic vRNPs in late-stage influenza virus infection. In addition, the PB2 protein of the polymerase complex was shown to form a direct interaction with the RAB11 protein.
Momose, F. et al. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS ONE 6, e21123 (2011).
Avilov, S. V., Moisy, D., Naffakh, N. & Cusack, S. Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine 30, 7411–7417 (2012).
Vale, R. D., Reese, T. S. & Sheetz, M. P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50 (1985).
Rietdorf, J. et al. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nature Cell Biol. 3, 992–1000 (2001).
Eisfeld, A. J., Kawakami, E., Watanabe, T., Neumann, G. & Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85, 6117–6126 (2011). This paper established a clear and tight relationship between RAB11 vesicle trafficking and the trafficking of nuclear-exported influenza vRNPs between the MTOC and the plasma membrane in late-stage infection. In addition, RAB11 expression and functional GTPase activity were shown to be crucial for influenza vRNP cytoplasmic transport.
Shaw, M. L., Stone, K. L., Colangelo, C. M., Gulcicek, E. E. & Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 4, e1000085 (2008).
Kawaguchi, A., Matsumoto, K. & Nagata, K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J. Virol. 86, 11086–11095 (2012).
de Lucas, S., Peredo, J., Marion, R. M., Sanchez, C. & Ortin, J. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. J. Virol. 84, 7603–7612 (2010).
Chaineau, M., Danglot, L., Proux-Gillardeaux, V. & Galli, T. Role of HRB in clathrin-dependent endocytosis. J. Biol. Chem. 283, 34365–34373 (2008).
Pryor, P. R. et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 (2008).
Eisfeld, A. J., Neumann, G. & Kawaoka, Y. Human immunodeficiency virus rev-binding protein is essential for influenza A virus replication and promotes genome trafficking in late-stage infection. J. Virol. 85, 9588–9598 (2011).
McCown, M. F. & Pekosz, A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79, 3595–3605 (2005).
Iwatsuki-Horimoto, K. et al. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80, 5233–5240 (2006).
Chen, B. J., Leser, G. P., Jackson, D. & Lamb, R. A. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82, 10059–10070 (2008).
Noda, T. et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439, 490–492 (2006). This paper was the first to describe the organization pattern of influenza vRNPs in virus particles and provided strong support for the selective vRNP packaging model.
Chou, Y. Y. et al. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl Acad. Sci. USA 109, 9101–9106 (2012).
Fujii, Y., Goto, H., Watanabe, T., Yoshida, T. & Kawaoka, Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl Acad. Sci. USA 100, 2002–2007 (2003). This paper was the first to identify segment-specific packaging signals for an influenza vRNP, which supported the selective incorporation model of vRNP packaging.
Watanabe, T., Watanabe, S., Noda, T., Fujii, Y. & Kawaoka, Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77, 10575–10583 (2003).
Ozawa, M. et al. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 81, 30–41 (2007).
Liang, Y., Hong, Y. & Parslow, T. G. cis-acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79, 10348–10355 (2005).
Liang, Y., Huang, T., Ly, H. & Parslow, T. G. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 82, 229–236 (2008).
Hutchinson, E. C., Curran, M. D., Read, E. K., Gog, J. R. & Digard, P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82, 11869–11879 (2008).
Gog, J. R. et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 35, 1897–1907 (2007).
Muramoto, Y. et al. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80, 2318–2325 (2006).
Marsh, G. A., Hatami, R. & Palese, P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81, 9727–9736 (2007).
Noda, T. et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nature Commun. 3, 639 (2012). This paper was the first to describe the three-dimensional structure of influenza vRNPs within virus particles. The data suggest that inter-segment vRNP–vRNP interactions occur within virions and that vRNPs are incorporated into progeny virus particles in a specific pattern.
Fournier, E. et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40, 2197–2209 (2012).
Fournier, E. et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 30, 7359–7367 (2012).
Gavazzi, C. et al. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic Acids Res. 41, 1241–1254 (2013).
Essere, B. et al. Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proc. Natl Acad. Sci. USA 110, E3840–E3848 (2013).
Gavazzi, C. et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc. Natl Acad. Sci. USA 110, 16604–16609 (2013). This paper provides an intriguing set of experiments that suggest the existence of a bona fide inter-segment vRNA–vRNA interaction that occurs in IAV-infected cells.
Acknowledgements
The authors are grateful to S. Watson for scientific editing of the manuscript. This work was supported by grants-in-aid from the Ministry of Health, Labour, and Welfare, Japan, by ERATO (Japan Science and Technology Agency) and by National Institute of Allergy and Infectious Diseases Public Health Service research grants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Eisfeld, A., Neumann, G. & Kawaoka, Y. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13, 28–41 (2015). https://doi.org/10.1038/nrmicro3367
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3367