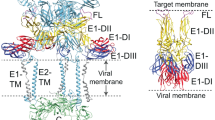
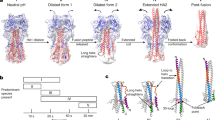
Key Points
-
Fusion proteins, which are viral envelope glycoproteins, participate actively in fusing viral and target-cell membranes.
-
Fusion proteins are thought to adopt a metastable conformation that relaxes to a highly stable conformation during the membrane-fusion event. This highly stable structure has been characterized for several viruses and has shown that there is a common structural motif.
-
For influenza viruses, and now also for paramyxoviruses, structural information is available for the metastable form, and this information indicates the nature of the protein conformational changes that accompany membrane fusion.
-
Although influenza viruses trigger fusion inside endosomes, for many viruses the fusion event occurs at the plasma membrane of the target cell and, in these cases, extra proteins in the viral envelope might be involved in assisting the fusion process.
-
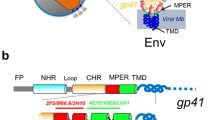
For human immunodeficiency virus (HIV), large conformational changes in the glycoprotein gp120 accompany receptor binding and these changes might be involved in priming the fusion protein gp41.
-
An increased understanding of the structural pathways that connect the metastable and highly stable conformers of the fusion protein is helping us to discover candidate drugs that block viral entry into cells.
Abstract
The fusion of viral membranes with target-cell membranes is an essential step in the entry of enveloped viruses into cells, and recent X-ray structures of paramyxoviral envelope proteins have provided new insights into protein-mediated plasma-membrane fusion. Here, we review our understanding of the structural transitions that are involved in this fusion pathway, compare it to our understanding of influenza virus membrane fusion, and discuss the implications for retroviral membrane fusion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Skehel, J. J. & Wiley, D. C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 (2000). A comprehensive review of the known properties of the influenza virus haemagglutinin and the structural basis of these properties.
Sollner, T., Bennett, M. K., Whiteheart, S. W., Scheller, R. H. & Rothman, J. E. A protein assembly–disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993).
Weissenhorn, W. et al. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16, 3–9 (1999).
Heinz, F. X. & Allison, S. L. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55, 231–269 (2000).
Crennell, S., Takimoto, T., Portner, A. & Taylor, G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nature Struct. Biol. 7, 1068–1074 (2000). This paper presents the structure of the HN attachment protein of NDV and provides a basis for interpreting data that link this protein to the fusion activity of the F protein.
Chen, L. et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9, 255–266 (2001). Together with influenza HA, the structure presented in this paper is one of only two that provide information on the pre-fusion conformation of a type I viral fusion protein.
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 (tm) resolution. Nature 289, 366–373 (1981).
Chen, J. et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95, 409–417 (1998).
Bullough, P. A., Hughson, F. M., Skehel, J. J. & Wiley, D. C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Chen, J., Skehel, J. J. & Wiley, D. C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl Acad. Sci. USA 96, 8967–8972 (1999).
Durrer, P. et al. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J. Biol. Chem. 271, 13417–13421 (1996).
Skehel, J. J. et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl Acad. Sci. USA 79, 968–972 (1982).
Kozlov, M. M. & Chernomordik, L. V. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys. J. 75, 1384–1396 (1998).
Bentz, J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 78, 886–900 (2000).
Gruenke, J. A., Armstrong, R. T., Newcomb, W. W., Brown, J. C. & White, J. M. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 76, 4456–4466 (2002).
Godley, L. et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68, 635–645 (1992).
Wharton, S. A., Skehel, J. J. & Wiley, D. C. Studies of influenza haemagglutinin-mediated membrane fusion. Virology 149, 27–35 (1986).
Ruigrok, R. W. H. et al. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 155, 484–497 (1986).
Stegmann, T. & Helenius, A. in Viral Fusion Mechanisms (ed. Bentz, J.) 89–111 (CRC Press, Boca Raton, Florida, USA, 1993).
Ward, C. W. & Dopheide, T. A. Influenza virus haemagglutinin. Structural predictions suggest that the fibrillar appearance is due to the presence of a coiled-coil. Aust. J. Biol. Sci. 33, 441–447 (1980).
Buckland, R. & Wild, F. Leucine zipper motif extends. Nature 338, 547 (1989).
Chambers, P., Pringle, C. R. & Easton, A. J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 71, 3075–3080 (1990).
Chen, L. et al. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology 290, 290–299 (2001).
Calder, L. J. et al. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271, 122–131 (2000).
Dutch, R. E., Hagglund, R. N., Nagel, M. A., Paterson, R. G. & Lamb, R. A. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281, 138–150 (2001).
Paterson, R. G., Russell, C. J. & Lamb, R. A. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270, 17–30 (2000).
Wharton, S. A., Skehel, J. J. & Wiley, D. C. Temperature dependence of fusion by Sendai virus. Virology 271, 71–78 (2000).
Lamb, R. A. Paramyxovirus fusion: a hypothesis for changes. Virology 197, 1–11 (1993).
Bousse, T., Takimoto, T., Gorman, W. L., Takahashi, T. & Portner, A. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204, 506–514 (1994).
Tanabayashi, K. & Compans, R. W. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. Virology 70, 6112–6118 (1996).
Stone-Hulslander, J. & Morrison, T. G. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73, 3630–3637 (1999).
Bousse, T., Takimoto, T. & Portner, A. A single amino acid change enhances the fusion promotion activity of human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Virology 209, 654–657 (1995).
Deng, R. et al. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253, 43–54 (1999).
Takimoto, T., Taylor, G. L., Connaris, H. C., Crennell, S. J. & Portner, A. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus–cell membrane fusion. J. Virol. 76, 13028–13033 (2002).
Yao, Q., Hu, X. & Compans, R. W. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71, 650–656 (1997).
Stone-Hulslander, J. & Morrison, T. G. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71, 6287–6295 (1997).
Earl, P. L., Broder, C. C., Doms, R. W. & Moss, B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71, 2674–2684 (1997).
Earl, P. L. et al. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68, 3015–3026 (1994).
Edinger, A. L. et al. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J. Virol. 73, 4062–4073 (1999).
Moore, J. P., McKeating, J. A., Weiss, R. A. & Sattentau, Q. J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250, 1130–1142 (1990).
Poignard, P., Fouts, T., Naniche, D., Moore, J. P. & Sattentau, Q. J. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183, 473–484 (1996).
Weissenhorn, W. et al. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc. Natl Acad. Sci. USA 94, 6065–6069 (1997).
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997).
Yang, Z. N. et al. The crystal structure of the SIV gp41 ectodomain at 1.47 (tm) resolution. J. Struct. Biol. 126, 131–144 (1999). This paper describes a high-resolution study of the fusion-core structure of simian immunodeficiency virus (SIV), which expands on the information that has been reported in parallel studies of HIV.
Thali, M., Furman, C., Helseth, E., Repke, H. & Sodroski, J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J. Virol. 66, 5516–5524 (1992).
Sattentau, Q. J. & Moore, J. P. Conformational changes induced in the human immunodeficiency virus envelope glycoproteins by soluble CD4 binding. J. Exp. Med. 174, 407–415 (1991).
Thali, M. et al. Characterization of human immunodeficiency virus type 1 (HIV-1) gp120 neutralization epitopes exposed upon gp120–CD4 binding. J. Virol. 67, 3978–3988 (1993).
Sattentau, Q. J., Moore, J. P., Vignaux, F., Traincard, F. & Poignard, P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67, 7383–7393 (1993).
Myszka, D. G. et al. Energetics of the HIV gp120–CD4 binding reaction. Proc. Natl Acad. Sci. USA 97, 9026–9031 (2000).
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002). This article describes a thermodynamic study of the binding of gp120 to CD4, which indicates that very large conformational changes accompany the binding event.
Binley, J. M. et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 (2000).
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997).
Kwong, P. D., Wyatt, R., Sattentau, Q. J., Sodroski, J. & Hendrickson, W. A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74, 1961–1972 (2000).
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Wild, C., Oas, T., McDanal, C., Bolognesi, D. & Matthews, T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl Acad. Sci. USA 89, 10537–10541 (1992).
Wild, C. T., Shugars, D. C., Greenwell, T. K., McDanal, C. B. & Matthews, T. J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl Acad. Sci. USA 91, 9770–9774 (1994).
Rapaport, D., Ovadia, M. & Shai, Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14, 5524–5531 (1995).
Lambert, D. M. et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl Acad. Sci. USA 93, 2186–2191 (1996).
Yao, Q. & Compans, R. W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223, 103–112 (1996).
Lu, M., Blacklow, S. C. & Kim, P. S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nature Struct. Biol. 2, 1075–1082 (1995).
Kilby, J. M. et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nature Med. 4, 1302–1307 (1998). This work describes early clinical data, which indicate that new medicines might emerge from our growing understanding of the molecular basis of viral membrane fusion.
Rimsky, L. T., Shugars, D. C. & Matthews, T. J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72, 986–993 (1998).
Chan, D. C., Chutkowski, C. T. & Kim, P. S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl Acad. Sci. USA 95, 15613–15617 (1998).
Eckert, D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99, 103–115 (1999).
Russell, C. J., Jardetzky, T. S. & Lamb, R. A. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20, 4024–4034 (2001). This paper decribes an important study of conformational changes in the F protein, which were probed using the inhibitory effects of N- and C-peptides.
Young, J. K., Li, D., Abramowitz, M. C. & Morrison, T. G. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 73, 5945–5956 (1999).
Furuta, R. A., Wild, C. T., Weng, Y. & Weiss, C. D. Capture of an early fusion-active conformation of HIV-1 gp41. Nature Struct. Biol. 5, 276–279 (1998).
Melikyan, G. B. et al. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151, 413–423 (2000).
Doms, R. W. & Moore, J. P. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151, F9–F14 (2000).
Koshiba, T. & Chan, D. C. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278, 7573–7579 (2003).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Meth. Enzymol. 277, 505–524 (1997).
Nicholls, A. GRASP: graphical representation and analysis of surface properties. Biophys. J. 64, A116 (1993).
Wild, T. F., Malvoisin, E. & Buckland, R. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 72, 439–442 (1991).
Ebata, S. N., Cote, M. -J., Yong Kang, C. & Dimock, K. The fusion and hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus 3 are both required for fusion. Virology 183, 437–441 (1991).
Hu, X., Ray, R. & Compans, R. W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66, 1528–1534 (1992).
Bagai, S. & Lamb, R. A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenxza virus 3 or Newcastle disease virus. J. Virol. 69, 6712–6719 (1995).
Ito, M. et al. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J. Virol. 71, 9855–9858 (1997).
Ito, M., Nishio, M., Komada, H., Ito, Y. & Tsurudome, M. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81, 719–727 (2000).
Tsurudome, M. et al. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 75, 8999–9009 (2001).
Sergel, T. A., McGinnes, L. W. & Morrison, T. G. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74, 5101–5107 (2000).
Seth, S., Vincent, A. & Compans, R. W. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77, 167–178 (2003).
McGinnes, L. W., Reitter, J. N., Gravel, K. & Morrison, T. G. Evidence for mixed membrane topology of the Newcastle disease virus fusion protein. J. Virol. 77, 1951–1963 (2003).
Wyde, P. R. et al. CL387626 exhibits marked and unusual antiviral activity against respiratory syncytial virus in tissue culture and in cotton rats. Antiviral Res. 38, 31–42 (1998).
Aulabaugh, A. et al. Inhibition of respiratory syncytial virus by a new class of chemotherapeutic agents. Drugs of the Future 25, 287–294 (2000).
Andries, K. et al. R170591, a new antiviral with picomolar activity against respiratory syncytial virus. Antiviral Res. 50, S1–S94 (2001).
Debnath, A. K., Radigan, L. & Jiang, S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 42, 3203–3209 (1999).
Jiang, S., Zhao, Q. & Debnath, A. K. Peptide and non-peptide HIV fusion inhibitors. Curr. Pharm. Des. 8, 563–580 (2002).
Zhou, G. et al. The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core. Bioorg. Med. Chem. 8, 2219–2227 (2000).
Smith, B. J., Lawrence, M. C. & Colman, P. M. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15, 365–371 (2002).
McKimm-Breschkin, J. L. et al. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob. Agents Chemother. 40, 40–46 (1996).
Blick, T. J. et al. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology 214, 475–484 (1995).
Colman, P. M. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 3, 1687–1696 (1994).
Greengard, O., Poltoratskaia, N., Leikina, E., Zimmerberg, J. & Moscona, A. The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J. Virol. 74, 11108–11114 (2000).
Neurath, A. R., Strick, N., Jiang, S., Li, Y. Y. & Debnath, A. K. Anti-HIV-1 activity of cellulose acetate phthalate: synergy with soluble CD4 and induction of 'dead-end' gp41 six-helix bundles. BMC Infect. Dis. 2, 6 (2002).
Baker, K. A., Dutch, R. E., Lamb, R. A. & Jardetzky, T. S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3, 309–319 (1999). This paper describes the structure of the complex of N- and C-peptides from the paramyxovirus F protein and links this family of viruses to HIV.
Zhao, X., Singh, M., Malashkevich, V. N. & Kim, P. S. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl Acad. Sci. USA 97, 14172–14177 (2000).
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Swiss-Prot
FURTHER INFORMATION
Glossary
- VIRAL FUSION PROTEIN
-
A glycoprotein that is anchored to the viral envelope. Its extracellular domain causes the fusion of the viral membrane with a membrane of target cells.
- METASTABLE STATE
-
A state that is marginally stable (in the case of fusion proteins, by virtue of the existence of a 'nearby' stable state that is separated from it by an energy barrier).
- HR-A AND HR-B
-
Heptad repeats or helical regions A and B of viral fusion proteins, which associate with one another in the post-fusion conformation.
- COILED-COILS
-
Two or more helical segments of polypeptide that align either parallel or anti-parallel to each other and then twist around one another in a rope-like fashion. Such an arrangement can often be predicted from amino-acid sequence analysis.
- PARAMYXOVIRUSES
-
The Paramyxoviridae family has two subfamilies — Paramyxovirinae (which includes parainfluenza viruses, mumps virus and measles virus) and Pneumovirinae (which includes respiratory syncytial virus). Members of both subfamilies have a related fusion protein.
- N-PEPTIDES AND C-PEPTIDES
-
Peptides that are derived from heptad repeat or helical region (HR)-A and HR-B of viral fusion proteins, respectively.
Rights and permissions
About this article
Cite this article
Colman, P., Lawrence, M. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol 4, 309–319 (2003). https://doi.org/10.1038/nrm1076
Issue Date:
DOI: https://doi.org/10.1038/nrm1076
This article is cited by
-
In Silico Studies of Phytoconstituents from Piper longum and Ocimum sanctum as ACE2 and TMRSS2 Inhibitors: Strategies to Combat COVID-19
Applied Biochemistry and Biotechnology (2023)
-
SARS-CoV fusion peptides induce membrane surface ordering and curvature
Scientific Reports (2016)
-
A generic screening platform for inhibitors of virus induced cell fusion using cellular electrical impedance
Scientific Reports (2016)
-
Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240
Scientific Reports (2016)
-
Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV
Nature Structural & Molecular Biology (2016)