Abstract
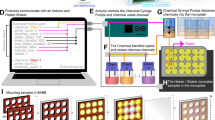
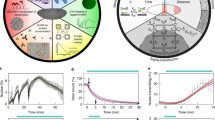
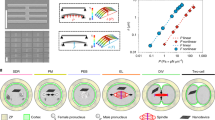
Spindle assembly and chromosome segregation rely on a complex interplay of biochemical and mechanical processes. Analysis of this interplay requires precise manipulation of endogenous cellular components and high-resolution visualization. Here we provide a protocol for generating an extract from individual Drosophila syncytial embryos that supports repeated mitotic nuclear divisions with native characteristics. In contrast to the large-scale, metaphase-arrested Xenopus egg extract system, this assay enables the serial generation of extracts from single embryos of a genetically tractable organism, and each extract contains dozens of autonomously dividing nuclei that can be prepared and imaged within 60–90 min after embryo collection. We describe the microscopy setup and micropipette production that facilitate single-embryo manipulation, the preparation of embryos and the steps for making functional extracts that allow time-lapse microscopy of mitotic divisions ex vivo. The assay enables a unique combination of genetic, biochemical, optical and mechanical manipulations of the mitotic machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gillespie, P.J., Gambus, A. & Blow, J.J. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin-associated proteins. Methods (2012).
Raschle, M. et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008).
Luo, X., Nerlick, S., An, W. & King, M.L. Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development 138, 589–598 (2011).
Rooke, N. & Underwood, J. In vitro RNA splicing in mammalian cell extracts. Curr. Protoc. Cell Biol. 11, 11.17–11.20 (2001).
Roca, X. & Karginov, F.V. RNA biology in a test tube-an overview of in vitro systems/assays. RNA 3, 509–527 (2012).
Hoskins, A.A. et al. Ordered and dynamic assembly of single spliceosomes. Science 331, 1289–1295 (2011).
Azzu, V. & Brand, M.D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 123, 578–585 (2010).
Jeske, M., Moritz, B., Anders, A. & Wahle, E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J. 30, 90–103 (2011).
Medenbach, J., Seiler, M. & Hentze, M.W. Translational control via protein-regulated upstream open reading frames. Cell 145, 902–913 (2011).
Shimamura, A. & Worcel, A. The assembly of regularly spaced nucleosomes in the Xenopus oocyte S-150 extract is accompanied by deacetylation of histone H4. J. Biol. Chem. 264, 14524–14530 (1989).
Tse, C., Sera, T., Wolffe, A.P. & Hansen, J.C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18, 4629–4638 (1998).
Azuma, Y. Analysis of SUMOylation of topoisomerase IIα with Xenopus egg extracts. Methods Mol. Biol. 582, 221–231 (2009).
Lohka, M.J. & Masui, Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 220, 719–721 (1983).
Lohka, M.J. & Maller, J.L. Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. J. Cell Biol. 101, 518–523 (1985).
Murray, A.W. & Kirschner, M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature 339, 275–280 (1989).
Hannak, E. & Heald, R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1, 2305–2314 (2006).
Kornbluth, S., Yang, J. & Powers, M. Analysis of the cell cycle using Xenopus egg extracts. Curr. Protoc. Cell Biol. 11.11.1–11.11.14 (2006).
Telley, I.A., Gaspar, I., Ephrussi, A. & Surrey, T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 197, 887–895 (2012).
Wuhr, M. et al. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 18, 1256–1261 (2008).
Holloway, S.L., Glotzer, M., King, R.W. & Murray, A.W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73, 1393–1402 (1993).
Shamu, C.E. & Murray, A.W. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 117, 921–934 (1992).
Murray, A.W., Desai, A.B. & Salmon, E.D. Real-time observation of anaphase in vitro. Proc. Natl. Acad. Sci. USA 93, 12327–12332 (1996).
Desai, A., Maddox, P.S., Mitchison, T.J. & Salmon, E.D. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J. Cell Biol. 141, 703–713 (1998).
Itabashi, T. et al. Probing the mechanical architecture of the vertebrate meiotic spindle. Nat. Methods 6, 167–172 (2009).
Shimamoto, Y., Maeda, Y.T., Ishiwata, S., Libchaber, A.J. & Kapoor, T.M. Insights into the micromechanical properties of the metaphase spindle. Cell 145, 1062–1074 (2011).
Brown, K.S. et al. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 176, 765–770 (2007).
Kieserman, E.K. & Heald, R. Mitotic chromosome size scaling in Xenopus. Cell Cycle 10, 3863–3870 (2011).
Loughlin, R., Wilbur, J.D., McNally, F.J., Nedelec, F.J. & Heald, R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407 (2011).
Bowes, J.B. et al. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 36, D761–767 (2008).
McCleland, M.L. & O'Farrell, P.H. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr. Biol. 18, 245–254 (2008).
Oliveira, R.A., Hamilton, R.S., Pauli, A., Davis, I. & Nasmyth, K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12, 185–192 (2010).
Cao, J., Crest, J., Fasulo, B. & Sullivan, W. Cortical actin dynamics facilitate early-stage centrosome separation. Curr. Biol. 20, 770–776 (2010).
Sharp, D.J. et al. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 11, 241–253 (2000).
Cheerambathur, D.K. et al. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 177, 995–1004 (2007).
Baker, J., Theurkauf, W.E. & Schubiger, G. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J. Cell Biol. 122, 113–121 (1993).
von Dassow, G. & Schubiger, G. How an actin network might cause fountain streaming and nuclear migration in the syncytial Drosophila embryo. J. Cell Biol. 127, 1637–1653 (1994).
Johansen, J. & Johansen, K.M. The spindle matrix through the cell cycle in Drosophila. Fly 3, 213–220 (2009).
Scott, M.P., Storti, R.V., Pardue, M.L. & Rich, A. Cell-free protein synthesis in lysates of Drosophila melanogaster cells. Biochemistry 18, 1588–1594 (1979).
Kunert, N. et al. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J. 28, 533–544 (2009).
Gebauer, F., Corona, D.F., Preiss, T., Becker, P.B. & Hentze, M.W. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5' and 3' UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 18, 6146–6154 (1999).
Dahmann, C. Drosophila: Methods and Protocols (Humana Press, 2008).
Pfeiffer, B.D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010).
Lund, V.K., DeLotto, Y. & DeLotto, R. A set of P-element transformation vectors permitting the simplified generation of fluorescent fusion proteins in Drosophila melanogaster. Fly 5, 255–260 (2011).
Lindsley, D.L. & Zimm, G.G. The genome of Drosophila melanogaster (Academic Press, 1992).
Bieling, P., Telley, I.A., Hentrich, C., Piehler, J. & Surrey, T. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 95, 555–580 (2010).
Schubiger, G. & Edgar, B. Using inhibitors to study embryogenesis. Methods Cell Biol. 44, 697–713 (1994).
Brust-Mascher, I. & Scholey, J.M. Microinjection techniques for studying mitosis in the Drosophila melanogaster syncytial embryo. J. Vis. Exp. 31, e1382 (2009).
Cold Spring Harbor Protocols. Drosophila apple juice-agar plates. Cold Spring Harb. Protoc. doi:10.1101/pdb.rec065672 (2011).
Brown, K.T. & Flaming, D.G. Advanced Micropipette Techniques for Cell Physiology (Wiley, 1986).
Foe, V.E. & Alberts, B.M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31–70 (1983).
Musacchio, A. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. B 366, 3595–3604 (2011).
Sigrist, S., Jacobs, H., Stratmann, R. & Lehner, C.F. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 14, 4827–4838 (1995).
Peti, W. & Page, R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 51, 1–10 (2007).
Telley, I.A., Bieling, P. & Surrey, T. Obstacles on the microtubule reduce the processivity of kinesin-1 in a minimal in vitro system and in cell extract. Biophys. J. 96, 3341–3353 (2009).
Edgar, B.A., Sprenger, F., Duronio, R.J., Leopold, P. & O'Farrell, P.H. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 8, 440–452 (1994).
Stiffler, L.A., Ji, J.Y., Trautmann, S., Trusty, C. & Schubiger, G. Cyclin A and B functions in the early Drosophila embryo. Development 126, 5505–5513 (1999).
Acknowledgements
We thank the EMBL Mechanical Workshop for equipment construction, and the Bloomington Stock Center for fly stocks. We thank E.M. Tranfield (EMBL) for suggesting correlative light-electron microscopy as an application. I.A.T. was a recipient of an Advanced Researcher Fellowship from the Swiss National Science Foundation (SNF), and acknowledges additional support from the EMBL. I.G. was a recipient of an EMBO long-term fellowship and an EMBL EIPOD fellowship. A.E. acknowledges support of the EMBL. T.S. acknowledges support of the EMBL and Cancer Research UK. This work was supported by the EMBL.
Author information
Authors and Affiliations
Contributions
I.A.T. and I.G. determined the correct timing of extraction. I.A.T. designed the instrument, drew up the protocol and performed the experiments. I.A.T. and T.S. conceived the cycle arrest experiment. I.A.T. prepared the figures. A.E. and T.S. supervised the work. All authors discussed the work and contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Telley, I., Gáspár, I., Ephrussi, A. et al. A single Drosophila embryo extract for the study of mitosis ex vivo. Nat Protoc 8, 310–324 (2013). https://doi.org/10.1038/nprot.2013.003
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.003
This article is cited by
-
Chromatin remodeling in Drosophila preblastodermic embryo extract
Scientific Reports (2018)
-
Systems and synthetic biology approaches in understanding biological oscillators
Quantitative Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.