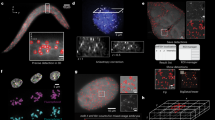
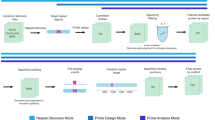
Abstract
Array painting is a technique that uses microarray technology to rapidly map chromosome translocation breakpoints. Previous methods to map translocation breakpoints have used fluorescence in situ hybridization (FISH) and have consequently been labor-intensive, time-consuming and restricted to the low breakpoint resolution imposed by the use of metaphase chromosomes. Array painting combines the isolation of derivative chromosomes (chromosomes with translocations) and high-resolution microarray analysis to refine the genomic location of translocation breakpoints in a single experiment. In this protocol, we describe array painting by isolation of derivative chromosomes using a MoFlo flow sorter, amplification of these derivatives using whole-genome amplification and hybridization onto commercially available oligonucleotide microarrays. Although the sorting of derivative chromosomes is a specialized procedure requiring sophisticated equipment, the amplification, labeling and hybridization of DNA is straightforward, robust and can be completed within 1 week. The protocol described produces good quality data; however, array painting is equally achievable using any combination of the available alternative methodologies for chromosome isolation, amplification and hybridization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Millar, J.K. et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423 (2000).
Gu, W., Zhang, F. & Lupski, J.R. Mechanisms for human genomic rearrangements. Pathogenetics 1, 4 (2008).
McMullan, T.W. et al. A candidate gene for congenital bilateral isolated ptosis identified by molecular analysis of a de novo balanced translocation. Hum. Genet. 110, 244–250 (2002).
van Bakel, I., Holt, S., Craig, I. & Boyd, Y. Sequence analysis of the breakpoint regions of an X;5 translocation in a female with Duchenne muscular dystrophy. Am. J. Hum. Genet. 57, 329–336 (1995).
Spitz, F. et al. A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics 79, 493–498 (2002).
Mills, K.I. et al. Amplification and sequencing of genomic breakpoints located within the M-bcr region by Vectorette-mediated polymerase chain reaction. Leukemia 6, 481–483 (1992).
Fiegler, H. et al. Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J. Med. Genet. 40, 664–670 (2003).
Pinkel, D. et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20, 207–211 (1998).
Gribble, S.M. et al. Ultra-high resolution array painting facilitates breakpoint sequencing. J. Med. Genet. 44, 51–58 (2007).
Baptista, J. et al. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am. J. Hum. Genet. 82, 927–936 (2008).
Fauth, C. et al. Micro-array analyses decipher exceptional complex familial chromosomal rearrangement. Hum. Genet. 119, 145–153 (2006).
Foster, R.E. et al. Characterization of a 3;6 translocation associated with renal cell carcinoma. Genes Chromosomes Cancer 46, 311–317 (2007).
Gribble, S.M. et al. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J. Med. Genet. 42, 8–16 (2005).
Rodriguez-Perales, S. et al. Cloning of a new familial t(3;8) translocation associated with conventional renal cell carcinoma reveals a 5 kb microdeletion and no gene involved in the rearrangement. Hum. Mol. Genet. 13, 983–990 (2004).
Vermeesch, J.R. et al. Tetrasomy 12pter-12p13.31 in a girl with partial Pallister–Killian syndrome phenotype. Eur. J. Med. Genet. 48, 319–327 (2005).
Backx, L. et al. Array painting using microdissected chromosomes to map chromosomal breakpoints. Cytogenet Genome Res. 116, 158–166 (2007).
Veltman, I.M. et al. Chromosomal breakpoint mapping by arrayCGH using flow-sorted chromosomes. Biotechniques 35, 1066–1070 (2003).
Gribble, S.M. et al. Applications of combined DNA microarray and chromosome sorting technologies. Chromosome Res. 12, 35–43 (2004).
Howarth, K.D. et al. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene 27, 3345–3359 (2008).
Carter, N.P. et al. Reverse chromosome painting: a method for the rapid analysis of aberrant chromosomes in clinical cytogenetics. J. Med. Genet. 29, 299–307 (1992).
Telenius, H. et al. Chromatid contamination can impair the purity of flow-sorted metaphase chromosomes. Cytometry 14, 97–101 (1993).
van den Engh, G., Trask, B., Lansdorp, P. & Gray, J. Improved resolution of flow cytometric measurements of Hoechst- and chromomycin-A3-stained human chromosomes after addition of citrate and sulfite. Cytometry 9, 266–270 (1988).
Mayall, B.H. et al. The DNA-based human karyotype. Cytometry 5, 376–385 (1984).
Freeman, J.L. et al. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genomics 8, 195 (2007).
Ng, B.L., Yang, F. & Carter, N.P. Flow analysis and sorting of microchromosomes (<3 Mb). Cytometry A 71, 410–413 (2007).
Gel electrophoresis. In Molecular Cloning, a Laboratory Manual, Vol. 1 (eds. Sambrook, J & Russell D.W.) chapter 54, p. 5.1–5.13 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Fiegler, H., Redon, R. & Carter, N.P. Construction and use of spotted large-insert clone DNA microarrays for the detection of genomic copy number changes. Nat. Protoc. 2, 577–587 (2007).
Acknowledgements
The labeling procedure described in Steps 39–43 was originally developed by Heike Fiegler28 and slightly modified for this protocol. We acknowledge Diana Rajan and Leong Siew Hong for proofreading and checking this protocol. We thank John Crolla (Wessex Regional Genetics Laboratory) for supplying the t(3;20) cell line used in Figure 3. This work was supported by the Wellcome Trust [Grant no. WT077008].
Author information
Authors and Affiliations
Contributions
S.M.G. and B.L.N. contributed equally in writing this paper; E.P. verified the protocol and proofread this paper; T.F. contributed to figures; N.P.C. originally developed this procedure and supervised this study.
Corresponding author
Rights and permissions
About this article
Cite this article
Gribble, S., Ng, B., Prigmore, E. et al. Array painting: a protocol for the rapid analysis of aberrant chromosomes using DNA microarrays. Nat Protoc 4, 1722–1736 (2009). https://doi.org/10.1038/nprot.2009.183
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.183
This article is cited by
-
Loss of H3K9 trimethylation alters chromosome compaction and transcription factor retention during mitosis
Nature Structural & Molecular Biology (2023)
-
Chromosome instability and aneuploidy in the mammalian brain
Chromosome Research (2023)
-
Comparative chromosome painting in Chacoan peccary, Catagonus wagneri
Journal of Applied Genetics (2021)
-
Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction
Nature Communications (2020)
-
A Comparative Study of Pygmy Hippopotamus (Choeropsis liberiensis) Karyotype by Cross-Species Chromosome Painting
Journal of Mammalian Evolution (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.