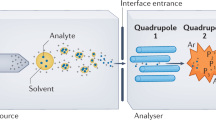
Abstract
We describe a matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (MS)-based assay for human papillomavirus (HPV) genotyping—the restriction fragment mass polymorphism (RFMP) assay, which is based on mass measurement of genotype-specific oligonucleotide fragments generated by TypeIIS restriction endonuclease cleavage after recognition sites have been introduced by PCR amplification. The use of a TypeIIS restriction enzyme makes the RFMP assay independent of sequence and applicable to a wide variety of HPV genotypes, because these enzymes have cleavage sites at a fixed distance from their recognition sites. After PCR amplification, samples are subjected to restriction enzyme digestion with FokI and BtsCI and desalting using Oasis purification plates, followed by analysis by MALDI-TOF MS. Overall, the protocol is simple, takes ∼4–4.5 h and can accurately detect and identify at least 74 different HPV genotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Muñoz, N. et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348, 518–527 (2003).
Chow, V.T., Loh, E., Yeo, W.M., Tan, S.Y. & Chan, R. Identification of multiple genital HPV types and sequence variants by consensus and nested type-specific PCR coupled with cycle sequencing. Pathology 32, 204–208 (2000).
Laconi, S. et al. One-step detection and genotyping of human papillomavirus in cervical samples by reverse hybridization. Diagn. Mol. Pathol. 10, 200–206 (2001).
Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. J. Natl. Cancer Inst. 92, 397–402 (2000).
Terry, G. et al. Detection of high-risk HPV types by the hybrid capture 2 test. J. Med. Virol. 65, 155–162 (2001).
Schiffman, M.H. et al. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85, 958–964 (1993).
Walboomers, J.M. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 (1999).
Bulkmans, N.W., Rozendaal, L., Voorhorst, F.J., Snijders, P.J. & Meijer, C.J. Long-term protective effect of high-risk human papillomavirus testing in population-based cervical screening. Br. J. Cancer 92, 1800–1802 (2005).
Clavel, C. et al. Negative human papillomavirus testing in normal smears selects a population at low risk for developing high-grade cervical lesions. Br. J. Cancer 90, 1803–1808 (2004).
Haff, L.A. & Smirnov, I.P. Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res. 7, 378–388 (1997).
Laken, S.J. et al. Genotyping by mass spectrometric analysis of short DNA fragments. Nat. Biotechnol. 16, 1352–1356 (1998).
Ross, P., Hall, L., Smirnov, I.P. & Haff, L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat. Biotechnol. 16, 1347–1351 (1998).
Abdi, F., Bradbury, E.M., Doggett, N. & Chen, X. Rapid characterization of DNA oligomers and genotyping of single nucleotide polymorphism using nucleotide-specific mass tags. Nucleic Acids Res. 29, 61 (2001).
Wolfe, J.L. et al. A genotyping strategy based on incorporation and cleavage of chemically modified nucleotides. Proc. Natl. Acad. Sci. USA 99, 11073–11078 (2002).
Mauger, F. et al. SNP genotyping using alkali cleavage of RNA/DNA chimeras and MALDI time-of-flight mass spectrometry. Nucleic Acids Res. 34, e18 (2006).
Sauer, S., Reinhardt, R., Lehrach, H. & Gut, I.G. Single-nucleotide polymorphisms: analysis by mass spectrometry. Nat. Protoc. 1, 1761–1771 (2006).
Stuyver, L. et al. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Microbiol. 38, 702–707 (2000).
Stuyver, L., Wyseur, A., van Arnhem, W., Hernandez, F. & Maertens, G. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34, 2259–2266 (1996).
Kim, H.S. et al. Evaluation of methods for monitoring drug resistance in chronic hepatitis B patients during lamivudine therapy based on mass spectrometry and reverse hybridization. Antivir. Ther. 10, 441–449 (2005).
Hong, S.P. et al. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J. Hepatol. 40, 837–844 (2004).
Kim, Y.J. et al. Population genotyping of hepatitis C virus by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of short DNA fragments. Clin. Chem. 51, 1123–1131 (2005).
Szybalski, W., Kim, S.C., Hasan, N. & Podhajska, A.J. Class-IIS restriction enzymes-a review. Gene 100, 13–26 (1991).
Lee, C.H. et al. Predominance of hepatitis B virus YMDD mutants is prognostic of viral DNA breakthrough. Gastroenterology 130, 1144–1152 (2006).
Yeon, J.E. et al. Resistance to adefovir dipivoxil in lamivudine-resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut 55, 1488–1495 (2006).
Paik, Y.H. et al. Clinical impact of early detection for YMDD mutant on the outcomes of long-term lamivudine therapy in patients with chronic hepatitis B. Antivir. Ther. 11, 447–455 (2006).
Lee, Y.S. et al. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology 43, 1385–1391 (2006).
Chung, H.J. et al. Comparison of efficacy of human papilloma virus genotyping assays using restriction fragment mass polymorphism and DNA chip analysis in patients with abnormal pap smear and uterine cervical cancer. Korean J. Pathol. 40, 439–347 (2006).
Lee, E.H. et al. Human papilloma virus genotyping assay using restriction fragment mass polymorphism analysis, and its comparison with sequencing and hybrid capture assays. Korean J. Lab. Med. 27, 62–68 (2007).
Ahn, S.H. et al. Association of genetic variations in CCR5 and its ligand, RANTES with clearance of hepatitis B virus in Korea. J. Med. Virol. 78, 1564–1571 (2006).
Hwang, S.H., Oh, H.B., Choi, S.E., Hong, S.P. & Yoo, W. Effective screening of informative single nucleotide polymorphisms using the novel method of restriction fragment mass polymorphism. J. Int. Med. Res. 35, 827–835 (2007).
Sohn, S.K. et al. Efficacy of dose escalation of imatinib mesylate in patients with cytogenetic or hematologic resistance. Leuk. Lymphoma 48, 1659–1661 (2007).
Smirnov, I.P., Hall, L.R., Ross, P.L. & Haff, L.A. Application of DNA-binding polymers for preparation of DNA for analysis by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1427–1432 (2001).
Little, D.P. et al. MALDI on a chip: analysis of arrays of low-femtomole to subfemtomole quantities of synthetic oligonucleotides and DNA diagnostic products by a piezoelectric pipet. Anal. Chem. 69, 4540–4546 (1997).
Shahgholi, M., Garcia, B.A., Chiu, N.H., Heaney, P.J. & Tang, K. Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved. Nucleic Acids Res. 29, e91 (2001).
Wu, K.J., Steding, A. & Becker, C.H. Matrix-assisted laser desorption time-of-flight mass spectrometry of oligonucleotides using 3-hydroxypicolinic acid as an ultraviolet-sensitive matrix. Rapid Commun. Mass Spectrom. 7, 142–146 (1993).
Kim, Y., Hurst, G.B., Doktycz, M.J. & Buchanan, M.V. Improving spot homogeneity by using polymer substrates in matrix-assisted laser desorption/ionization mass spectrometry of oligonucleotides. Anal. Chem. 73, 2617–2624 (2001).
Gravitt, P.E. et al. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38, 357–361 (2000).
Kwok, S. & Higuchi, R. Avoiding false positives with PCR. Nature 339, 237–238 (1989).
Wright, T.C. et al. 2001 Consensus Guidelines for the management of women with cervical cytological abnormalities. JAMA 287, 2120–2129 (2002).
Fontaine, V. et al. Evaluation of combined general primer-mediated PCR sequencing and type-specific PCR strategies for determination of human papillomavirus genotypes in cervical cell specimens. J. Clin. Microbiol. 45, 928–934 (2007).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Hong, S., Shin, SK., Lee, E. et al. High-resolution human papillomavirus genotyping by MALDI-TOF mass spectrometry. Nat Protoc 3, 1476–1484 (2008). https://doi.org/10.1038/nprot.2008.136
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.136
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.