Abstract
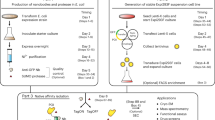
The present purification protocol applies to target proteins that are fused to a double tag, such as NusA–His6, through a linker that includes a protease-recognition sequence. It involves two steps of immobilized metal ion affinity chromatography (IMAC). NusA stabilizes the passenger protein during translation, whereas the His-tag enables affinity purification of the fusion. The eluate resulting from the first IMAC is buffer-exchanged to remove the imidazole and to achieve optimal conditions for the enzymatic cleavage performed by a His-tagged recombinant protease. The digested sample is loaded directly for a second IMAC step and the target protein is selectively recovered in the flow-through. The resin binds residual non-digested fusion protein, double-tagged moiety, protease and any contaminant that bound the affinity resin and was eluted from the first IMAC. The purity of the target protein usually makes a further purification step unnecessary for most of the lab applications. It takes less than 5 hours to purify the protein from a 5 g pellet.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Waugh, D.S. Making the most of affinity tags. Trends Biotechnol. 23, 316–320 (2005).
Davis, G.D., Elisee, C., Newham, D.M. & Harrison, R.G. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65, 382–388 (1999).
De Marco, V., Stier, G., Blandin, S. & de Marco, A. The solubility and stability of recombinant proteins is increased by their fusion to NusA. Biochem. Biophys. Res. Comm. 322, 766–771 (2004).
Saluta, M. & Bell, P.A. Troubleshooting GST fusion protein expression in E. coli. Life Sci. News 1, 1–3 (1998).
Kaplan, W. et al. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: a detoxification enzyme and fusion-protein affinity tag. Protein Sci. 6, 399–406 (1997).
Nilsson, J. et al. Multiple affinity domains for the detection, purification and immobilization of recombinant proteins. J. Mol. Recognit. 9, 585–594 (1996).
Pryor, K.D. & Leiting, B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding protein double-affinity fusion system. Protein Expr. Purif. 10, 309–319 (1997).
Smith, P.A. et al. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 26, 1414–1420 (1998).
Nallamsetty, S., Austin, B.P., Penrose, K.J. & Waugh, D.S. Gateway vectors for the production of combinatorially-tagged His6–MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 14, 1435–1442 (2005).
Cabrita, L.D., Dai, W. & Bottomley, S.P. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 6, 12 (2006).
Dümmler, A., Lawrence, A.-M. & de Marco, A. Simplified screening for the detection of soluble constructs expressed in E. coli using a modular set of vectors. Microbial Cell Factories 4, 34 (2005).
Nominé, Y. et al. A strategy for optimizing the monodispersity of the fusion proteins: application to purification of recombinant HPV E6 oncoprotein. Prot. Eng. 14, 297–305 (2001).
de Marco, A., Vigh, L., Diamant, S. & Goloubinoff, P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol over-expressed molecular chaperones. Cell Stress Chaperones 10, 329–339 (2005).
Kapust, R.B. et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
de Marco, A. Two-step metal affinity purification of double-tagged (NusA–His6) fusion proteins. Nat Protoc 1, 1538–1543 (2006). https://doi.org/10.1038/nprot.2006.289
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.289
This article is cited by
-
Structural and functional analysis of target recognition by the lymphocyte adaptor protein LNK
Nature Communications (2021)
-
Protocol for preparing proteins with improved solubility by co-expressing with molecular chaperones in Escherichia coli
Nature Protocols (2007)
-
The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins
Nature Protocols (2007)
-
Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies
Molecular and Cellular Biochemistry (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.