Abstract
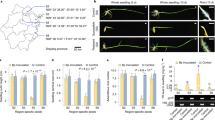
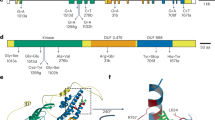
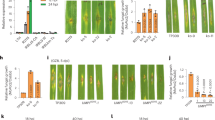
The major disease resistance gene Xa4 confers race-specific durable resistance against Xanthomonas oryzae pv. oryzae, which causes the most damaging bacterial disease in rice worldwide. Although Xa4 has been one of the most widely exploited resistance genes in rice production worldwide, its molecular nature remains unknown. Here we show that Xa4, encoding a cell wall-associated kinase, improves multiple traits of agronomic importance without compromising grain yield by strengthening the cell wall via promoting cellulose synthesis and suppressing cell wall loosening. Strengthening of the cell wall by Xa4 enhances resistance to bacterial infection, and also increases mechanical strength of the culm with slightly reduced plant height, which may improve lodging resistance of the rice plant. The simultaneous improvement of multiple agronomic traits conferred by Xa4 may account for its widespread and lasting utilization in rice breeding programmes globally.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Johnson, R. Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology 71, 567–568 (1981).
Kou, Y. & Wang, S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185 (2010).
Zhang, H. & Wang, S. Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195 (2013).
Kou, Y. & Wang, S. Toward an understanding of the molecular basis of quantitative disease resistance in rice. J. Biotechnol. 159, 283–290 (2012).
Peng, J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999).
Sasaki, A. et al. Green revolution: a mutant gibberellin synthesis gene in rice. Nature 416, 701–702 (2002).
Zhang, B. & Zhou, Y. Rice brittleness mutants: a way to open the ‘black box’ of monocot cell wall biosynthesis. J. Integr. Plant Biol. 53, 136–142 (2011).
Ogawa, T., Tabien, R. E., Busto, G. A., Khush, G. S. & Mew, T. W. The relationships between Xa-3, Xa-4, and Xa-4b for resistances to rice bacterial blight. Rice Genet. Newslet. 3, 83–84 (1986).
Leach, J. E., Vera Cruz, C. M., Bai, J. & Leung, H. Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224 (2001).
Webb, K. M., Garcia, E., Vera Cruz, C. M. & Leach, J. E. Influence of rice development on the function of bacterial blight resistance genes. Eur. J. Plant Pathol. 128, 399–407 (2010).
Khush, G. S. & Virk, P. S. IR Varieties and Their Impact 8–32 (International Rice Research Institute, 2005).
Arif, M. et al. Identification of bacterial blight resistance genes Xa4 in Pakistani rice germplasm using PCR. African J. Biotechnol. 7, 541–545 (2008).
Zhang, Q. Genetic and improvement of resistance to bacterial blight of hybrid rice in China. Rice Science. 16, 83–92 (2009).
Sun, X., Yang, Z., Wang, S. & Zhang, Q. Identification of a 47kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor. Appl. Genet. 106, 683–687 (2003).
Sun, X. et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encoding a LRR receptor kinase-like protein. Plant J. 37, 517–527 (2004).
Xiang, Y., Cao, Y., Xu, C., Li, X. & Wang, S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor. Appl. Genet. 113, 1347–1355 (2006).
Sun, X., Cao, Y. & Wang, S. Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiol. 140, 998–1008 (2006).
Li, Z. K. et al. A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet. 261, 58–63 (1999).
Li, Z. K. et al. Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc. Natl Acad. Sci. USA 103, 7994–7999 (2006).
Chen, H., Wang, S. & Zhang, Q. New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology 92, 750–754 (2002).
Cao, Y. et al. The expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177, 523–533 (2007).
Somerville, C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22, 53–78 (2006).
Wang, L. et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282–297 (2010).
Cosgrove, D. J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–886 (2005).
Ding, X. et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 20, 228–240 (2008).
Fu, J. et al. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155, 589–602 (2011).
Tao, Z. et al. A pair of allelic WRKY genes play opposite role in rice-bacteria interactions. Plant Physiol. 151, 936–948 (2009).
Shen, X. et al. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 64, 86–99 (2010).
Deng, H., Liu, H., Li, X., Xiao, J. & Wang, S. A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 158, 876–889 (2012).
Liu, H., Li, X., Xiao, J. & Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8, 2 (2012).
Ke, Y., Liu, H., Li, X., Xiao, J. & Wang, S. Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host − pathogen interactions. Plant J. 78, 619–631 (2014).
Wakuta, S. et al. OsJAR1 and OsJAR2 are jasmonyl-L-isoleucine synthases involved in wound- and pathogen-induced jasmonic acid signalling. Biochem. Biophys. Res. Commun. 409, 634–639 (2011).
Zhang, S. B. et al. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139, 1107–1124 (2005).
Diener, A. C. & Ausubel, F. M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321 (2005).
Li, H., Zhou, S. Y., Zhao, W. S., Su, S. H. & Peng, Y. L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69, 337–346 (2009).
Zuo, W. et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157 (2015).
Hurni, S. et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl Acad. Sci. USA 112, 8780–8785 (2015).
Wolf, S., Hématy, K. & Höfte, H. Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63, 381–407 (2012).
Padmavati, M., Santhivel, N., Thara, K. V. & Reddy, A. R. Differential sensitivity of rice pathogens to growth inhibition by flavonoids. Phytochemistry 46, 449–502 (1997).
Riemann, M. et al. Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238 (2013).
Shimizu, T. et al. OsJAR1 contributes mainly to biosynthesis of the stress-induced jasmonoyl-isoleucine involved in defense responses in rice. Biosci. Biotechnol. Biochem. 77, 1556–1564 (2013).
González, J. F. et al. A proteomic study of Xanthomonas oryzae pv. oryzae in rice xylem sap. J. Proteomics 75, 5911–5919 (2012).
Ellis, C., Karafyllidis, I., Wasternack, C. & Turner, J. G. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566 (2002).
Ko, J. H., Kim, J. H., Jayanty, S. S., Howe, G. & Han, K. H. Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J. Exp. Bot. 57, 2923–2936 (2006).
Sharma, R. et al. Transcriptional dynamics during cell wall removal and regeneration reveals key genes involved in cell wall development in rice. Plant Mol. Biol. 77, 391–406 (2011).
Yan, C., Yan, S., Zeng, X., Zhang, Z. & Gu, M. Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J. Genet. Genomics 34, 1019–1127 (2007).
Ge, X., Chu, Z., Lin, Y. & Wang, S. A tissue culture system for different germplasms of indica rice. Plant Cell Rep. 25, 392–402 (2006).
Chen, J. et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl Acad. Sci. USA 105, 11436–11441 (2008).
Qiu, D. et al. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20, 492–499 (2007).
Acknowledgements
We thank Y. Zhou of the Institute of Genetics and Developmental Biology, Chinese Academy of Science and M. Gu of the Yangzhou University for providing rice cesa4 mutant seeds, M. Xu of the China Agricultural University for providing the ZmWAK construct and L. Peng of the Huazhong Agricultural University for helping with the analysis of the cell wall composition. We also thank the RIKEN Yokohama Institute for providing CesA4 (J023093O20), CesA7 (J023009D02) and CesA9 (J023081B08) cDNA clones. This work was supported by grants from the National Natural Science Foundation of China (31330062), the National Key Research and Development Program of China (2016YFD0100903), and the National Program of High Technology Development of China (2014AA10A600).
Author information
Authors and Affiliations
Contributions
K.H. designed and performed most of the experiments, analysed the data and drafted the manuscript; J.C., J.Z., F.X., Y.K., H.Z., W.X., H.L., Y.Cui., Y.Cao. and X.S. helped to perform histology, electron microscopy, protein subcellular localization, DNA hybridization and metabolite analyses and to generate some transgenic rice plants; J.X., X.L. and Q.Z. provided biochemical and molecular analysis support and field management; S.W. supervised the project, designed some of the experiments, interpreted data and revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–23, Supplementary Tables 1–6. (PDF 2658 kb)
Rights and permissions
About this article
Cite this article
Hu, K., Cao, J., Zhang, J. et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nature Plants 3, 17009 (2017). https://doi.org/10.1038/nplants.2017.9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.9
This article is cited by
-
The ZmWAKL–ZmWIK–ZmBLK1–ZmRBOH4 module provides quantitative resistance to gray leaf spot in maize
Nature Genetics (2024)
-
Comparative genomics analysis of WAK/WAKL family in Rosaceae identify candidate WAKs involved in the resistance to Botrytis cinerea
BMC Genomics (2023)
-
Genome-wide identification and expression analysis of wall-associated kinase (WAK) and WAK-like kinase gene family in response to tomato yellow leaf curl virus infection in Nicotiana benthamiana
BMC Plant Biology (2023)
-
Genome-wide characterization of the wall-associated kinase-like (WAKL) family in sesame (Sesamum indicum) identifies a SiWAKL6 gene involved in resistance to Macrophomina Phaseolina
BMC Plant Biology (2023)
-
TALE-induced immunity against the bacterial blight pathogen Xanthomonas oryzae pv. oryzae in rice
Phytopathology Research (2022)