Abstract
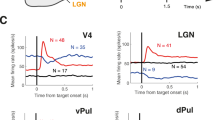
Neural responses are typically characterized by computing the mean firing rate, but response variability can exist across trials. Many studies have examined the effect of a stimulus on the mean response, but few have examined the effect on response variability. We measured neural variability in 13 extracellularly recorded datasets and one intracellularly recorded dataset from seven areas spanning the four cortical lobes in monkeys and cats. In every case, stimulus onset caused a decline in neural variability. This occurred even when the stimulus produced little change in mean firing rate. The variability decline was observed in membrane potential recordings, in the spiking of individual neurons and in correlated spiking variability measured with implanted 96-electrode arrays. The variability decline was observed for all stimuli tested, regardless of whether the animal was awake, behaving or anaesthetized. This widespread variability decline suggests a rather general property of cortex, that its state is stabilized by an input.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Briggman, K.L., Abarbanel, H.D. & Kristan, W.B. Jr. Optical imaging of neuronal populations during decision-making. Science 307, 896–901 (2005).
Arieli, A., Sterkin, A., Grinvald, A. & Aertsen, A. Dynamics of ongoing activity: explanation of the large variability in evoked cortical responses. Science 273, 1868–1871 (1996).
Monier, C., Chavane, F., Baudot, P., Graham, L.J. & Fregnac, Y. Orientation and direction selectivity of synaptic inputs in visual cortical neurons: a diversity of combinations produces spike tuning. Neuron 37, 663–680 (2003).
Finn, I.M., Priebe, N.J. & Ferster, D. The emergence of contrast-invariant orientation tuning in simple cells of cat visual cortex. Neuron 54, 137–152 (2007).
Kohn, A., Zandvakili, A. & Smith, M.A. Correlations and brain states: from electrophysiology to functional imaging. Curr. Opin. Neurobiol. 19, 434–438 (2009).
Azouz, R. & Gray, C.M. Cellular mechanisms contributing to response variability of cortical neurons in vivo. J. Neurosci. 19, 2209–2223 (1999).
Fiser, J., Chiu, C. & Weliky, M. Small modulation of ongoing cortical dynamics by sensory input during natural vision. Nature 431, 573–578 (2004).
Kisley, M.A. & Gerstein, G.L. Trial-to-trial variability and state-dependent modulation of auditory-evoked responses in cortex. J. Neurosci. 19, 10451–10460 (1999).
Churchland, M.M., Yu, B.M., Ryu, S.I., Santhanam, G. & Shenoy, K.V. Neural variability in premotor cortex provides a signature of motor preparation. J. Neurosci. 26, 3697–3712 (2006).
Rickert, J., Riehle, A., Aertsen, A., Rotter, S. & Nawrot, M.P. Dynamic encoding of movement direction in motor cortical neurons. J. Neurosci. 29, 13870–13882 (2009).
Sussillo, D. & Abbott, L.F. Generating coherent patterns of activity from chaotic neural networks. Neuron 63, 544–557 (2009).
Abbott, L.F., Rajan, K. & Sompolinsky, H. Interactions between intrinsic and stimulus-dependent activity in recurrent neural networks. in Neuronal Variability and its Functional Significance (eds Ding, M. & Glanzman, D.) (in the press).
Poulet, J.F. & Petersen, C.C. Internal brain state regulates membrane potential synchrony in barrel cortex of behaving mice. Nature 454, 881–885 (2008).
Shadlen, M.N. & Newsome, W.T. The variable discharge of cortical neurons: implications for connectivity, computation, and information coding. J. Neurosci. 18, 3870–3896 (1998).
van Vreeswijk, C. & Sompolinsky, H. Chaos in neuronal networks with balanced excitatory and inhibitory activity. Science 274, 1724–1726 (1996).
Mainen, Z.F. & Sejnowski, T.J. Reliability of spike timing in neocortical neurons. Science 268, 1503–1506 (1995).
Carandini, M. Amplification of trial-to-trial response variability by neurons in visual cortex. PLoS Biol. 2, e264 (2004).
Tolhurst, D.J., Movshon, J.A. & Dean, A.F. The statistical reliability of signals in single neurons in cat and monkey visual cortex. Vision Res. 23, 775–785 (1983).
Gur, M., Beylin, A. & Snodderly, D.M. Response variability of neurons in primary visual cortex (V1) of alert monkeys. J. Neurosci. 17, 2914–2920 (1997).
Nawrot, M.P. et al. Measurement of variability dynamics in cortical spike trains. J. Neurosci. Methods 169, 374–390 (2008).
Mitchell, J.F., Sundberg, K.A. & Reynolds, J.H. Differential attention-dependent response modulation across cell classes in macaque visual area V4. Neuron 55, 131–141 (2007).
Roweis, S. & Ghahramani, Z. A unifying review of linear gaussian models. Neural Comput. 11, 305–345 (1999).
Everitt, B.S. An Introduction to Latent Variable Models (Chapman & Hall, London, 1984).
Smith, M.A. & Kohn, A. Spatial and temporal scales of neuronal correlation in primary visual cortex. J. Neurosci. 28, 12591–12603 (2008).
Churchland, M.M., Afshar, A. & Shenoy, K.V. A central source of movement variability. Neuron 52, 1085–1096 (2006).
Yu, B.M. et al. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J. Neurophysiol. 102, 614–635 (2009).
Monier, C., Fournier, J. & Fregnac, Y. In vitro and in vivo measures of evoked excitatory and inhibitory conductance dynamics in sensory cortices. J. Neurosci. Methods 169, 323–365 (2008).
Britten, K.H., Newsome, W.T., Shadlen, M.N., Celebrini, S. & Movshon, J.A. A relationship between behavioral choice and the visual responses of neurons in macaque MT. Vis. Neurosci. 13, 87–100 (1996).
Horwitz, G.D. & Newsome, W.T. Target selection for saccadic eye movements: prelude activity in the superior colliculus during a direction-discrimination task. J. Neurophysiol. 86, 2543–2558 (2001).
Cohen, M.R. & Maunsell, J.H. Attention improves performance primarily by reducing interneuronal correlations. Nat. Neurosci. 12, 1594–1600 (2009).
Mandelblat-Cerf, Y., Paz, R. & Vaadia, E. Trial-to-trial variability of single cells in motor cortices is dynamically modified during visuomotor adaptation. J. Neurosci. 29, 15053–15062 (2009).
Kao, M.H., Doupe, A.J. & Brainard, M.S. Contributions of an avian basal ganglia-forebrain circuit to real-time modulation of song. Nature 433, 638–643 (2005).
Oram, M.W., Hatsopoulos, N.G., Richmond, B.J. & Donoghue, J.P. Excess synchrony in motor cortical neurons provides redundant direction information with that from coarse temporal measures. J. Neurophysiol. 86, 1700–1716 (2001).
Kara, P., Reinagel, P. & Reid, R.C. Low response variability in simultaneously recorded retinal, thalamic, and cortical neurons. Neuron 27, 635–646 (2000).
Osborne, L.C., Bialek, W. & Lisberger, S.G. Time course of information about motion direction in visual area MT of macaque monkeys. J. Neurosci. 24, 3210–3222 (2004).
Lee, D. & Seo, H. Neural and behavioral variability related to stochastic choices during a mixed-strategy game. in Neuronal Variability and its Functional Significance (eds Ding, M. & Glanzman, D.) (in the press).
Fortier, P.A., Smith, A.M. & Kalaska, J.F. Comparison of cerebellar and motor cortex activity during reaching: directional tuning and response variability. J. Neurophysiol. 69, 1136–1149 (1993).
Cohen, J.Y. et al. Difficulty of visual search modulates neuronal interactions and response variability in the frontal eye field. J. Neurophysiol. 98, 2580–2587 (2007).
Nauhaus, I., Busse, L., Carandini, M. & Ringach, D.L. Stimulus contrast modulates functional connectivity in visual cortex. Nat. Neurosci. 12, 70–76 (2009).
Werner, G. & Mountcastle, V.B. The variability of central neural activity in a sensory system and its implications for the central reflection of sensory events. J. Neurophysiol. 26, 958–977 (1963).
Wang, X.J. Decision making in recurrent neuronal circuits. Neuron 60, 215–234 (2008).
Churchland, M.M., Yu, B.M., Sahani, M. & Shenoy, K.V. Techniques for extracting single-trial activity patterns from large-scale neural recordings. Curr. Opin. Neurobiol. 17, 609–618 (2007).
Sugrue, L.P., Corrado, G.S. & Newsome, W.T. Matching behavior and the representation of value in the parietal cortex. Science 304, 1782–1787 (2004).
Cohen, M.R. & Newsome, W.T. Context-dependent changes in functional circuitry in visual area MT. Neuron 60, 161–173 (2008).
Armstrong, K.M., Fitzgerald, J.K. & Moore, T. Changes in visual receptive fields with microstimulation of frontal cortex. Neuron 50, 791–798 (2006).
Armstrong, K.M. & Moore, T. Rapid enhancement of visual cortical response discriminability by microstimulation of the frontal eye field. Proc. Natl. Acad. Sci. USA 104, 9499–9504 (2007).
Chang, S.W., Dickinson, A.R. & Snyder, L.H. Limb-specific representation for reaching in the posterior parietal cortex. J. Neurosci. 28, 6128–6140 (2008).
Priebe, N.J., Churchland, M.M. & Lisberger, S.G. Constraints on the source of short-term motion adaptation in macaque area MT. I. The role of input and intrinsic mechanisms. J. Neurophysiol. 88, 354–369 (2002).
Boch, R. & Fischer, B. Saccadic reaction times and activation of the prelunate cortex: parallel observations in trained rhesus monkeys. Exp. Brain Res. 50, 201–210 (1983).
Kihlberg, J.K., Herson, J.H. & Schotz, W.E. Square root transformation revisited. Appl. Stat. 21, 76–81 (1972).
Acknowledgements
This work was supported by a Helen Hay Whitney postdoctoral fellowship (M.M.C.), Burroughs Welcome Fund Career Awards in the Biomedical Sciences (M.M.C. and K.V.S.), Gatsby Charitable Foundation (M.S. and B.M.Y.), US National Institutes of Health (NIH), National Institute of Neurological Disorders and Stroke Collaborative Research in Computational Neuroscience grant R01-NS054283 (K.V.S. and M.S.), the Michael Flynn Stanford Graduate Fellowship (J.P.C.), the Howard Hughes Medical Institute and NIH grant EY05603 (W.T.N., L.P.S., M.R.C. and G.S.C.), a Howard Hughes Medical Institute predoctoral fellowship (M.R.C.), NIH grant EY014924 (T.M. and K.M.A.), Sloan Foundation (T.M. and K.V.S.), Pew Charitable Trust (T.M.), NIH EY015958 and EY018894 (M.A.S.), NIH EY02017 and EY04440 (J.A.M.), NIH EY016774 (A.K.), NIH 1 EY13138-01 (D.C.B., A.M.C., P.H. and B.B.S.), NIH EY019288 (N.J.P.), the Pew Charitable Trust (N.J.P.), EY04726 (D.F.), US National Defense Science and Engineering Graduate Fellowships (B.M.Y. and G.S.), National Science Foundation Graduate Research Fellowships (B.M.Y. and G.S.), the Christopher and Dana Reeve Foundation (K.V.S. and S.I.R.), and the awards from Stanford Center for Integrated Systems, National Science Foundation Center for Neuromorphic Systems Engineering at Caltech, Office of Naval Research, NIH Director's Pioneer Award 1DP1OD006409 and Whitaker Foundation (K.V.S.).
Author information
Authors and Affiliations
Contributions
M.M.C. wrote the manuscript, performed the Fano factor and factor analyses and created the figures. GPFA was developed by B.M.Y., J.P.C., M.S. and K.V.S. This application of factor analysis was devised by M.M.C. and B.M.Y. The mean-matched Fano factor was developed by M.M.C. and K.V.S. The conception for the study arose from conversations between M.M.C., K.V.S., B.M.Y., D.C.B., M.R.C., W.T.N. and J.A.M. V1 data (extracellular) were collected in the laboratory of J.A.M. by M.A.S. and A.K. and in the laboratory of A.K. V4 data were collected in the laboratory of T.M. by K.M.A. MT (plaid) data were collected in the laboratory of D.C.B. by A.M.C., P.H. and B.B.S. MT (dots) data were collected in the laboratory of W.T.N. by M.R.C. LIP and OFC data were collected in the laboratory of W.T.N. by L.P.S. using an experimental design developed by L.P.S. and G.S.C. PRR data were collected in the laboratory of L.H.S. by S.W.C. PMd data were collected in the laboratory of K.V.S. by B.M.Y., S.I.R., G.S. and M.M.C. MT (direction/area and speed) data were collected by N.J.P. and M.M.C. in the laboratory of S.G.L. Intracellularly recorded V1 data were collected by N.J.P. and I.M.F. in the laboratory of D.F. All authors contributed to manuscript revisions and editing, particularly J.A.M., W.T.N., L.P.S., D.F., J.P.C., B.M.Y. and K.V.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Notes 1–3 (PDF 1369 kb)
Supplementary Video 1
A movie version of Figure 7a. Data are from PMd, and show the decline in across-trial variance after the onset of the stimulus (a reach target). The movie spans 750 ms, beginning 400 ms before stimulus onset and ending 350 ms after. The movie ends before the go cue is given. Each black dot shows the state of PMd on one trial. Fifteen randomly-chosen trials are shown. Dots turn blue for a brief moment at the time of stimulus onset. Note the subsequent drop in the variance of the dot locations (i.e., a drop in firing-rate variance). This feature of the response is at least as clear as the change in mean dot location (i.e., the change in mean firing rates). G20040123 dataset. (AVI 488 kb)
Supplementary Video 2
As in Supplementary Video 1, but more time is shown and the trajectory of the RT-outlier trial is now included (red). The movie spans ∼1500 ms. This time-span differs slightly across trials, as they have different go-cue and movement-onset times. At the time of the go cue, each dot turns green and further progress is halted. Progress resumes once all trials have passed the time of their respective go cues. This re-aligns the data to the go cue, much as is commonly done in PSTH's. Traces end at movement onset. (AVI 1235 kb)
Supplementary Video 3
As in Supplementary Video 1, but for the G20040122 PMd dataset (that shown in Figure 7c). (AVI 505 kb)
Supplementary Video 4
As in Supplementary Video 2, but for the G20040122 PMd dataset (that shown in Figure 7c). (AVI 1280 kb)
Rights and permissions
About this article
Cite this article
Churchland, M., Yu, B., Cunningham, J. et al. Stimulus onset quenches neural variability: a widespread cortical phenomenon. Nat Neurosci 13, 369–378 (2010). https://doi.org/10.1038/nn.2501
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2501
This article is cited by
-
Activation of M1 cholinergic receptors in mouse somatosensory cortex enhances information processing and detection behaviour
Communications Biology (2024)
-
Co-dependent excitatory and inhibitory plasticity accounts for quick, stable and long-lasting memories in biological networks
Nature Neuroscience (2024)
-
Beyond expectations: residual dynamic mode decomposition and variance for stochastic dynamical systems
Nonlinear Dynamics (2024)
-
Correlated variability in primate superior colliculus depends on functional class
Communications Biology (2023)
-
Continuous multiplexed population representations of task context in the mouse primary visual cortex
Nature Communications (2023)