Abstract
The ribose of RNA nucleotides can be 2′-O-methylated (Nm). Despite advances in high-throughput detection, the inert chemical nature of Nm still limits sensitivity and precludes mapping in mRNA. We leveraged the differential reactivity of 2′-O-methylated and 2′-hydroxylated nucleosides to periodate oxidation to develop Nm-seq, a sensitive method for transcriptome-wide mapping of Nm with base precision. Nm-seq uncovered thousands of Nm sites in human mRNA with features suggesting functional roles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
Gene Expression Omnibus
Change history
28 February 2018
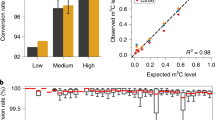
Nat. Methods 14, 695–698 (2017); published online 15 May 2017; corrected after print 28 February 2018 We were alerted by readers that the reported Nm consensus sequence in mRNA matches the 3′-adaptor sequence used in sequencing library preparations, and this could be caused by mispriming1. In our approach, the majority of RNA fragments without Nm at the 3′ end are blocked from ligating to the 3′ adaptor because of the presence of 3′ phosphate from the last oxidation elimination step (OE) (Fig. 1a of the original paper), while Nm sites accumulate at the 3′ ends (Supplementary Figs. 1 and 2; Supplementary Notes 1 and 2). However, because of the low Nm abundance in messenger RNA (mRNA), only very limited amounts of mRNA fragments carry 3′ Nm and thus can be successfully ligated to the 3′ adaptor. Mispriming could occur if the 3′ end of the reverse transcription (RT) primer hybridizes to a few bases of the 5′-ligated RNA (Fig. 1a). Although our method effectively identifies Nm sites in abundant ribosomal RNA (rRNA, Supplementary Fig. 1), its application to less abundant mRNA can be contaminated by mispriming, leading to false-positive Nm sites and the erroneous AGAUC motif on mRNA (original Fig. 2d), which corresponds to the 5′-end sequence of the 3′ adaptor.
References
Zhao, B.S., Roundtree, I.A. & He, C. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Machnicka,, M.A. et al. MODOMICS: a database of RNA modification pathways-2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Daffis, S. et al. Nature 468, 452–456 (2010).
Deryusheva, S., Choleza, M., Barbarossa, A., Gall, J.G. & Bordonné, R. RNA 18, 31–36 (2012).
Jöckel, S. et al. J. Exp. Med. 209, 235–241 (2012).
Liang, X.H., Liu, Q. & Fournier, M.J. RNA 15, 1716–1728 (2009).
Lin, J. et al. Nature 469, 559–563 (2011).
Somme, J. et al. RNA 20, 1257–1271 (2014).
Züst, R. et al. Nat. Immunol. 12, 137–143 (2011).
Shubina, M.Y., Musinova, Y.R. & Sheval, E.V. Biochemistry (Mosc.) 81, 941–950 (2016).
Byszewska, M., S´mietan´ski, M., Purta, E. & Bujnicki, J.M. RNA Biol. 11, 1597–1607 (2014).
Lacoux, C. et al. Nucleic Acids Res. 40, 4086–4096 (2012).
Kumar, S., Mapa, K. & Maiti, S. Biochemistry 53, 1607–1615 (2014).
Yildirim, I., Kierzek, E., Kierzek, R. & Schatz, G.C. J. Phys. Chem. B 118, 14177–14187 (2014).
Maden, B.E., Corbett, M.E., Heeney, P.A., Pugh, K. & Ajuh, P.M. Biochimie 77, 22–29 (1995).
Incarnato, D. et al. Nucleic Acids Res. 45, 1433–1441 (2017).
Birkedal, U. et al. Angew. Chem. Int. Ed. Engl. 127, 461–455 (2015).
Krogh, N. et al. Nucleic Acids Res. 44, 7884–7895 (2016).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Nucleic Acids Res. 44, e135 (2016).
Gumienny, R. et al. Nucleic Acids Res. 45, 2341–2353 http://dx.doi.org/10.1093/nar/gkw1321 (2017).
Hoernes, T.P. et al. Nucleic Acids Res. 44, 852–862 (2016).
Sonenberg, N. & Shatkin, A.J. Proc. Natl. Acad. Sci. USA 74, 4288–4292 (1977).
Weith, H.L. & Gilham, P.T. Science 166, 1004–1005 (1969).
Weith, H.L. & Gilham, P.T. J. Am. Chem. Soc. 89, 5473–5474 (1967).
Yu, B. et al. Science 307, 932–935 (2005).
Heinz, S. et al. Mol. Cell 38, 576–589 (2010).
Dai, Q. et al. Nm-seq protocol. Protocol Exchange http://dx.doi.org/10.1038/protex.2017.088 (2017).
Dominissini, D. et al. Nature 530, 441–446 (2016).
Kellner, S. et al. Nucleic Acids Res. 42, e142 (2014).
Langmead, B. & Salzberg, S.L. Nat. Methods 9, 357–359 (2012).
Martin, M. EMBnet Journal 17, 10–12 (2011).
Anders, S., Pyl, P.T. & Huber, W. Bioinformatics 31, 166–169 (2015).
Wang, K., Li, M. & Hakonarson, H. Nucleic Acids Res. 38, e164 (2010).
Zhou, W. et al. Nat. Methods 12, 1002–1003 (2015).
Kishore, S. et al. Genome Biol. 14, R45 (2013).
Quinlan, A.R. & Hall, I.M. Bioinformatics 26, 841–842 (2010).
Berkowitz, N.D. et al. BMC Bioinformatics 17, 215 (2016).
Acknowledgements
This work was supported by the US National Institutes of Health NHGRI RM1 HG008935 to C.H.; a grant from the Kahn Family Foundation to D.D. and G.R.; and grants from the Ernest and Bonnie Beutler Research Program, Flight Attendant Medical Research Institute (FAMRI) and the Israeli Centers of Excellence (I-CORE) Program (ISF grants no. 41/11 and no. 1796/12) to G.R. C.H. is an investigator of the Howard Hughes Medical Institute (HHMI). G.R. is a member of the Sagol Neuroscience Network and holds the Djerassi Chair for Oncology at the Sackler Faculty of Medicine, Tel-Aviv University, Israel. D.D. was supported by a Human Frontier Science Program (HFSP) long-term fellowship. Q.D. is supported by the National Institutes of Health grant K01 HG006699. We wish to thank M. Salmon-Divon for advice and help with bioinformatic analysis and R. Mashiach for help with chemical structure drawings.
Author information
Authors and Affiliations
Contributions
D.D., Q.D. and C.H. conceived the approach. D.D. and Q.D. developed the methods and performed experiments. D.D., S.M.-M., D.H. and N.K. analyzed the data. D.D., S.M.-M., N.A., G.R. and C.H. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Detection of Nm sites in HeLa rRNA
(a-b), Nm-seq profiles of human 18S (a) and 28S (b) rRNA above MCC-determined optimal threshold (blue). Known Nm sites are shown as red bars below. (c-d), Receiver Operating Characteristic (ROC, orange) and Mathews Correlation Coefficient (MCC, green) curves for human 18S (c) and 28S (d) rRNA plotted using increasing normalized 3′ end coverage thresholds at each position.
Supplementary Figure 2 Examples of Nm sites in HeLa mRNA.
Nm-seq plots of methylated transcripts: (a) NKIRAS1 (b) KLHL5. Normalized summed sequence coverage of Nm-seq and input are shown below and above the transcript, respectively. Individual paired-end reads within the Nm site window are shown in magnification.
Supplementary Figure 3 Features of the HeLa Nm methylome
(a) Distribution of 2′-O-methyl sites between the four nucleobases in the various transcript segments and overall. (b) Fraction of Nm sites detected within mRNA and ncRNA. (c) The percentage of methylated genes according to the number of Nm sites per gene. (d) The percentage of methylated genes increases with expression level.
Supplementary Figure 4 RNA secondary structure surrounding Nm sites, m6Am in mRNA and Gene Ontology (GO) analysis
The secondary structures of a 200-nt window centered on Nm sites was analyzed using the Structure Surfer tool based on: (a) PARS score (b) ds/ssRNA score and (c) DMS-seq. (d) LC-MS/MS quantification of internal (i.e., excluding the first transcribed nucleotide) m6A and internal m6Am in HeLa mRNA. The level of each modified nucleoside is presented as a percentage of the unmodified one. Mean values ± s.e.m. are shown, n = 3. (e) GO analysis of Nm-methylated HeLa genes relative to all adequately expressed genes (above the 1st quartile) reveals enrichment of GO terms related to cell-cell interactions, splicing and more (fold enrichment ≥ 2, Bonferroni corrected P ≤ 0.005). Fold-enrichment and P values are indicated for each category.
Supplementary Figure 5 Distribution of Nm sites in HeLa mRNA
(a) Distribution of Nm sites between exons, introns and alternatively spliced regions. (b) Metagene profile of Nm site distribution along a normalized mRNA transcript. (c) Metagene profile of Nm sites distribution relative to the first and nearest splice sites in a 400-nt non-normalized window. (d) Metagene profile of Nm site distribution along a normalized intron.
Supplementary Figure 6 Features of the Nm methylomes in HEK293 cells (part 1)
(a) Distribution of 2′-O-methyl sites between the four nucleobases in the various transcript segments and overall. (b) Fraction of Nm sites detected within mRNA and ncRNA. (c) Metagene profile of Nm sites distribution along a normalized mRNA transcript illustrated below. (d) Sequence logo of the most enriched motif identified by HOMER in 58.7% of all HEK293 Nm sites. (e) The percentage of methylated genes according to the number of Nm sites per gene.
Supplementary Figure 7 Features of the Nm methylomes in HEK293 cells (part 2)
(a) HEK293 Nm sites in different transcript segments of coding genes. (b) Distribution of Nm sites between exons, introns and alternatively spliced regions. (c) Metagene profile of Nm sites distribution relative to the first (blue) and nearest (red) splice sites in a 400-nt non-normalized window. (d) Distribution of Nm sites between the three codon positions. (e) Distribution of Nm sites among different amino acid codons.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–2 and Supplementary Notes 1–5.
Supplementary Data
Supplementary Data Set.
Supplementary Protocol
Supplementary Protocol 1.
Rights and permissions
About this article
Cite this article
Dai, Q., Moshitch-Moshkovitz, S., Han, D. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods 14, 695–698 (2017). https://doi.org/10.1038/nmeth.4294
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4294
This article is cited by
-
Epitranscriptomic modifications in mesenchymal stem cell differentiation: advances, mechanistic insights, and beyond
Cell Death & Differentiation (2024)
-
Small RNA modifications: regulatory molecules and potential applications
Journal of Hematology & Oncology (2023)
-
A patient-derived organoid-based study identified an ASO targeting SNORD14E for endometrial cancer through reducing aberrant FOXM1 Expression and β-catenin nuclear accumulation
Journal of Experimental & Clinical Cancer Research (2023)
-
MePMe-seq: antibody-free simultaneous m6A and m5C mapping in mRNA by metabolic propargyl labeling and sequencing
Nature Communications (2023)
-
A two-residue nascent-strand steric gate controls synthesis of 2′-O-methyl- and 2′-O-(2-methoxyethyl)-RNA
Nature Chemistry (2023)