Abstract
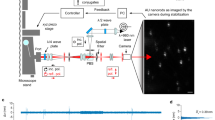
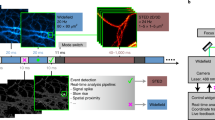
Electro-optical scanning (>1,000 frames/s) with pixel dwell times on the order of the lifetime of the fluorescent molecular state renders stimulated emission depletion (STED) nanoscopy temporally stochastic. Photon detection from a molecule occurs stochastically in one of several scanning frames, and the spatial origin of the photon is known with subdiffraction precision. Images are built up by binning consecutive frames, making the time resolution freely adjustable. We demonstrated nanoscopy of vesicle motions in living Drosophila larvae and the cellular uptake of viral particles with 5- to 10-ms temporal resolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hell, S.W. Science 316, 1153–1158 (2007).
Betzig, E. et al. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P.K. & Mason, M.D. Biophys. J. 91, 4258–4272 (2006).
Rust, M.J., Bates, M. & Zhuang, X.W. Nat. Methods 3, 793–795 (2006).
Sharonov, A. & Hochstrasser, R.M. Proc. Natl. Acad. Sci. USA 103, 18911–18916 (2006).
Egner, A. et al. Biophys. J. 93, 3285–3290 (2007).
Biteen, J.S. et al. Nat. Methods 5, 947–949 (2008).
Fölling, J. et al. Nat. Methods 5, 943–945 (2008).
Hell, S.W. & Wichmann, J. Opt. Lett. 19, 780–782 (1994).
Tsien, R.Y. & Backsai, B.J. in Handbook of Biological Confocal Microscopy 2nd edn. (ed. Pawley, J.) 459–478 (Plenum Press, New York, 1996).
Westphal, V. et al. Science 320, 246–249 (2008).
Huang, F. et al. Nat. Methods 10, 653–658 (2013).
Lauterbach, M.A., Ullal, C.K., Westphal, V. & Hell, S.W. Langmuir 26, 14400–14404 (2010).
Wu, X., Toro, L., Stefani, E. & Wu, Y. J. Microsc. 257, 31–38 (2015).
Rietdorf, J. & Stelzer, E.H.K. in Handbook of Biological Confocal Microscopy 3rd edn. (ed. Pawley, J.) Ch. 3, 43–58 (Springer, New York, 2006).
Chen, X., Leischner, U., Rochefort, N.L., Nelken, I. & Konnerth, A. Nature 475, 501–505 (2011).
Kremer, Y. et al. Opt. Express 16, 10066–10076 (2008).
Salomé, R. et al. J. Neurosci. Methods 154, 161–174 (2006).
Engelhardt, J., Özcelik, J. & Hell, S.W. Elektrooptische Anordnung zum ultraschnellen Abtasten eines Objektes. German patent DE 202010004547 U1 (2011) and European patent application WO2012171999 A1 (2012).
Diaspro, A., Chirico, G., Usai, C., Ramoino, P. & Dobrucki, J. in Handbook of Biological Confocal Microscopy 3rd edn. (ed. Pawley, J.) Ch. 39, 690–700 (Springer, New York, 2006).
Eggeling, C., Widengren, J., Rigler, R. & Seidel, C.A.M. Anal. Chem. 70, 2651–2659 (1998).
Tsien, R.Y., Ernst, L. & Waggoner, A. in Handbook of Biological Confocal Microscopy 3rd edn. (ed. Pawley, J.) Ch. 16, 338–352 (Springer, New York, 2006).
Donnert, G., Eggeling, C. & Hell, S.W. Nat. Methods 4, 81–86 (2007).
Bingen, P., Reuss, M., Engelhardt, J. & Hell, S.W. Opt. Express 19, 23716–23726 (2011).
Reuss, M., Engelhardt, J. & Hell, S.W. Opt. Express 18, 1049–1058 (2010).
Vogelsang, J. et al. Angew. Chem. Int. Ed. 47, 5465–5469 (2008).
Acknowledgements
We thank D. Richardson (MPI-BPC, now at Harvard University) and C. Gregor (MPI-BPC) for providing the TAT-EGFP samples, as well as H. Ta for advice on single-emitter STED measurements. We thank T. Hope (University of Illinois) for the plasmid pEGFP.Vpr. This work was supported by a Leibniz Award of the Deutsche Forschungsgemeinschaft (to S.W.H.) and by the German Federal Ministry of Education and Research (BMBF) within the project STEDlight (FKZ: 13N11173).
Author information
Authors and Affiliations
Contributions
J.S. and J.E. developed the EOD scanning system and set up the microscope. J.S. and J.Z. performed the experiments and analyzed the data. M.M., S.J. Sigrist, J.C. and H.-G.K. provided samples. J.M. performed single-antibody imaging. J.S., J.E., S.J. Sahl and S.W.H. wrote the manuscript. The latter also conceived and guided the project and is responsible for its main points. All authors discussed the data and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Increased photon yield for several fluorophores by ultrafast scanning compared to the conventional slow-scanning mode.
More than 40 comparative measurement series were performed in order to assess to which extent our fast scanner reduced bleaching. Samples with different labeling were studied with the different available laser configurations. For a single measurement, areas of the same size and of originally similar brightness within the same sample were scanned fast and slow respectively. The total image acquisition duration was kept the same for both cases to ensure the same light dose over the area. The pixel dwell time was 6.25 ns for the ultrafast scanning and 20 µs for the slow-scanning modality. Fluorophore molecules gradually bleached both in ultrafast- and slow-scanned areas, but the photon yield per applied light dose in ultrafast-scanned areas was clearly higher. (a-c) Examples of three measurement series on samples expressing yellow fluorescent protein (YFP): (a) labeled mitochondriae in fixed cells, subjected to continuous-wave (CW) excitation. (b) living neurons (cytosolic expression) in brain slices, subjected to pulsed excitation. (c) fixed neurons (cytosolic expression), subjected to pulsed excitation and CW-STED. The left axis shows the mean value of detected photons in the scanned areas after each consecutive scan. After each measurement series, the mean number of photons collected in the fast scanned area was divided by the mean number of photons in the equivalent slow-scanned area. High peak power of pulsed lasers presumably caused a significant amount of bleaching directly from the excited state before the molecules could cross to the triplet state. Thus, allowing triplet relaxation had a smaller effect. The obtained values are shown as green triangles (right axis of the graphs). These signal increase factors over many different analyzed samples are summarized in (d) with the photon yield in slow scanning given as 1.
Supplementary Figure 2 Resolution in ultrafast scanning STED nanoscopy.
Confocal and STED images of (a) yellow green fluorescent beads with a diameter of 24 nm, (b) MreB-rsEGFP in eisosomes in living E. coli, and (c) single-strand DNA labeled with the dye YOYO acquired by electro-optic temporally stochastic STED recordings. The line profiles indicate the resolution performance in the ultrafast scanning setup.
Supplementary Figure 3 Temporally stochastic STED imaging of single antibodies conjugated to Atto 647N.
(a) Examples of raw data and (b) data smoothed with a Gaussian filter (width of 2 pixels). Scale bars: 300 nm. (c) Histogram of numbers of detected signal photons per antibody for n=62 separate single-antibody scans. (d) Histogram of full-widths at half-maximum (FWHM) inferred by fitting symmetric Gaussian functions (with variable additive offset) to the smoothed data, for the same 62 single antibodies. (e) Number of signal photons vs. FWHM. The data show a mean resolution of ~58 nm for a mean of ~38 photons per antibody detected, at a background of ~0.06 photons per pixel (or ~0.6 photons per FHWM area). These data were recorded with fluorescence excitation at 640 nm (8 μW in back focal plane, 20 MHz) and STED at 775 nm (37.5 mW in back focal plane, 20 MHz) using a Leica 100× 1.46 NA oil immersion objective. (Pixel size: 17 nm, 30 000 frames of 5 ns dwell time, i.e. 150 μs effective pixel dwell time).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 786 kb)
Image build-up with ultra-fast scanning
This video shows how images of static samples are acquired. The sample with EGFP-labeled HI-Viruses is scanned with a line frequency of 250 kHz. The photons collected in each frame are accumulated online. One can see both the single frames and the cumulative image during data acquisition in STED as well as in confocal mode. When a sufficient number of photons are collected or when the number of photons per frame decreases, for example due to bleaching, the acquisition can be stopped manually. The video is displayed in real time and is repeated three times. (AVI 11537 kb)
Imaging of diffusing beads
Moving yellow green beads with a diameter of 100 nm are imaged with a frame rate of 793 fps. Many beads were diffusing freely in the water/glycerol film between the cover slip and the cover slide. Some beads were attached to the cover slip. The video in the upper-left panel shows the raw data during STED acquisition. A Gaussian filter with a full width at half maximum of 90 nm was applied to the data shown on the upper-right. The bottom-left panel depicts the raw data of the diffraction-limited confocal imaging of the same area. These data were smoothed with a Gaussian filter of 180 nm width and shown on the bottom-right. The video is slowed down 10 times for display. The original acquisition time was 1.66 sec. (AVI 19927 kb)
Imaging of vesicle dynamics in living Drosophila larvae
Video acquisition of vesicle trafficking in living Drosophila larvae with an effective frame rate of 125 fps through online binning (original scanning rate corresponds to a frame rate of 500 fps). The dynamics of neuropeptidergic vesicles were investigated, which carry EGFP-labeled proANF neuropeptide. Raw data is shown. (AVI 2555 kb)
Imaging of vesicle dynamics in living Drosophila larvae (laterally smoothed)
The raw data of Supplementary Video 3 is laterally smoothed with a Gaussian of 100 nm width and temporally smoothed over two frames. (AVI 3026 kb)
Imaging of HI-Virus uptake in living cells
Video acquisition of dynamics of EGFP-labeled HI-Viruses taken up by living HeLa cells. The effective frame rate of the acquisition is 125 fps. The data shown are laterally smoothed with a Gaussian of 100 nm and temporally smoothed over five frames. (AVI 1958 kb)
Source data
Rights and permissions
About this article
Cite this article
Schneider, J., Zahn, J., Maglione, M. et al. Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics. Nat Methods 12, 827–830 (2015). https://doi.org/10.1038/nmeth.3481
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3481
This article is cited by
-
Deep learning enables fast, gentle STED microscopy
Communications Biology (2023)
-
MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope
Nature Communications (2021)
-
Resolution-enhanced OCT and expanded framework of information capacity and resolution in coherent imaging
Scientific Reports (2021)
-
Converting lateral scanning into axial focusing to speed up three-dimensional microscopy
Light: Science & Applications (2020)
-
Real-time 3D single molecule tracking
Nature Communications (2020)