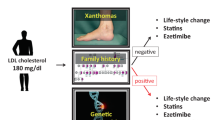
Abstract
The Haplotype Genetic Map (HapMap) is an invaluable resource to the cardiovascular researcher, enabling a decrease in cost and an increase in the efficiency and speed of discoveries in the laboratory. As cardiologists, we need to understand the vocabulary of genomics because the translation of scientific findings using HapMap could provide insight for improved care and therapeutic guidance of our patients. Genomics is the evaluation of genes as a dynamic system, in which genes interact to influence biologic pathways, networks and physiology. The HapMap promises to increase the efficiency of genomics in identifying cardiovascular-disease-related genes that could become vital for choosing relevant tests and providing preventative and curative therapies. In this Review, the HapMap will be described, to provide insight into the relevance of this work to cardiovascular practice, to clinical research in cardiovascular disease and to future discoveries in diagnostic and therapeutic modalities.
Key Points
-
The recently published Haplotype Genetic Map (HapMap) has the potential to increase the efficiency and speed, yet decrease the cost, of genetic discoveries
-
The HapMap is a catalogue of common haplotypes (i.e. a group of genetic variants, or genotypes closely linked on one chromosome and inherited as a unit), which is being used to find genes associated with human disease
-
Only by elucidating the complex interactions of multiple genetic variants, which are weak individually but that together confer a deleterious phenotype, will we begin to understand and effectively treat or prevent processes such as in-stent restenosis and subacute thrombosis
-
In the future, patients' genomes could be sequenced and incorporated into their medical record; an individual's genome sequence will then be vital in choosing tests as well as providing preventative and curative therapies
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Altshuler D et al. (2005) A haplotype map of the human genome. Nature 437: 1299–1320
Schmidt CW (2003) HapMap: building a database with blocks. EHP Toxicogenomics 111: A16
The International HapMap Consortium (2003) The International HapMap Project. Nature 426: 789–796
Thorisson GA et al. (2005) The International HapMap Project web site. Genome Res 15: 1592–1593
The International HapMap Consortium (2004) Integrating ethics and science in the International HapMap Project. Nat Rev Genet 5: 467–475
Kastrati A et al. (2006) Predictive factors of restenosis after coronary implantation of sirolimus- or paclitaxel-eluting stents. Circulation 113: 2293–2300
Di Castelnuovo A et al. (2005) Platelet glycoprotein IIb/IIIa polymorphism and coronary artery disease: implications for clinical practice. Am J Pharmacogenomics 5: 93–99
Koch W et al. (2004) Apolipoprotein E gene polymorphisms and thrombosis and restenosis after coronary artery stenting. J Lipid Res 45: 2221–2226
Koch W et al. (2004) Peroxisome proliferator-activated receptor gamma gene polymorphisms and restenosis in diabetic patients after stenting in coronary arteries. Diabetologia 47: 1126–1127
Angiolillo DJ et al. (2004) 807 C/T Polymorphism of the glycoprotein Ia gene and pharmacogenetic modulation of platelet response to dual antiplatelet treatment. Blood Coagul Fibrinolysis 15: 427–433
Angiolillo DJ et al. (2004) High clopidogrel loading dose during coronary stenting: effects on drug response and interindividual variability. Eur Heart J 25: 1903–1910
Angiolillo DJ et al. (2004) PlA polymorphism and platelet reactivity following clopidogrel loading dose in patients undergoing coronary stent implantation. Blood Coagul Fibrinolysis 15: 89–93
Angiolillo DJ et al. (2004) Platelet aggregation according to body mass index in patients undergoing coronary stenting: should clopidogrel loading-dose be weight adjusted? J Invasive Cardiol 16: 169–174
Chen SN et al. (2005) A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol 45: 1611–1619
Bezzina CR et al. (2006) Common sodium channel promoter haplotype in Asian subjects underlies variability in cardiac conduction. Circulation 113: 338–344
Anderson JL et al. (2000) A common variant of the AMPD1 gene predicts improved cardiovascular survival in patients with coronary artery disease. J Am Coll Cardiol 36: 1248–1252
Hingorani AD et al. (1999) A common variant of the endothelial nitric oxide synthase (Glu298Asp) is a major risk factor for coronary artery disease in the UK. Circulation 100: 1515–1520
Doris PA (2002) Hypertension genetics, single nucleotide polymorphisms, and the common disease:common variant hypothesis. Hypertension 39: 323–331
Samuels ME et al. (2001) Identification of a common variant in the lipoprotein lipase gene in a large Utah kindred ascertained for coronary heart disease: the -93G/D9N variant predisposes to low HDL-C/high triglycerides. Clin Genet 59: 88–98
Pritchard JK and Cox NJ (2002) The allelic architecture of human disease genes: common disease-common variant...or not? Hum Mol Genet 11: 2417–2423
Lunetta KL et al. (2004) Screening large-scale association study data: exploiting interactions using random forests. BMC Genet 5: 32
de Bakker PI et al. (2005) Efficiency and power in genetic association studies. Nat Genet 37: 1217–1223
Lander ES et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921
ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640
Waterston RH et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562
Stoneking M (2001) Single nucleotide polymorphisms: from the evolutionary past. Nature 409: 821–822
The International HapMap Project [http://www.hapmap.org]
Agema WR et al. (2002) A meta-analysis of the angiotensin-converting enzyme gene polymorphism and restenosis after percutaneous transluminal coronary revascularization: evidence for publication bias. Am Heart J 144: 760–768
Amant C et al. (1997) D allele of the angiotensin I-converting enzyme is a major risk factor for restenosis after coronary stenting. Circulation 96: 56–60
Rakugi H et al. (1994) Induction of angiotensin converting enzyme in the neointima after vascular injury. Possible role in restenosis. J Clin Invest 93: 339–346
Wijpkema JS et al. (2006) Restenosis after percutaneous coronary intervention is associated with the angiotensin-II type-1 receptor 1166A/C polymorphism but not with polymorphisms of angiotensin-converting enzyme, angiotensin-II receptor, angiotensinogen or heme oxygenase-1. Pharmacogenet Genomics 16: 331–337
Koch W et al. (2006) Association of transforming growth factor-β1 gene polymorphisms with myocardial infarction in patients with angiographically proven coronary heart disease. Arterioscler Thromb Vasc Biol 26: 1114–1119
Koch WJ (2004) Genetic and phenotypic targeting of beta-adrenergic signaling in heart failure. Mol Cell Biochem 263: 5–9
Faxon DP et al. (1984) Determinants of successful percutaneous transluminal coronary angioplasty: report from the National Heart, Lung, and Blood Institute Registry. Am Heart J 108: 1019–1023
Holmes DR Jr et al. (1984) Restenosis after percutaneous transluminal coronary angioplasty (PTCA): a report from the PTCA Registry of the National Heart, Lung, and Blood Institute. Am J Cardiol 53: 77C–81C
Venter JC et al. (2001) The sequence of the human genome. Science 291: 1304–1351
Goldstein DB and Weale ME (2001) Population genomics: linkage disequilibrium holds the key. Curr Biol 11: R576–R579
Smith AV et al. (2005) Sequence features in regions of weak and strong linkage disequilibrium. Genome Res 15: 1519–1534
Hinds DA et al. (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079
Nielsen R et al. (2005) Genomic scans for selective sweeps using SNP data. Genome Res 15: 1566–1575
Weir BS et al. (2005) Measures of human population structure show heterogeneity among genomic regions. Genome Res 15: 1468–1476
Forouhi NG and Sattar N (2006) CVD risk factors and ethnicity—a homogeneous relationship? Atheroscler 7 (Suppl): 11–19
Bertoni AG et al. (2006) Diabetic cardiomyopathy and subclinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis (MESA). Diabetes Care 29: 588–594
McCluskey J and Peh CA (1999) The human leucocyte antigens and clinical medicine: an overview. Rev Immunogenet 1: 3–20
Pearce E et al. (2005) Haplotype effect of the matrix metalloproteinase-1 gene on risk of myocardial infarction. Circ Res 97: 1070–1076
Kruger WD et al. (2000) Polymorphisms in the CBS gene associated with decreased risk of coronary artery disease and increased responsiveness to total homocysteine lowering by folic acid. Mol Genet Metab 70: 53–60
Wang Y et al. (2006) VKORC1 haplotypes are associated with arterial vascular diseases (stroke, coronary heart disease, and aortic dissection). Circulation 113: 1615–1621
Weinshilboum R (2003) Inheritance and drug response. N Engl J Med 348: 529–537
Ganesh SK et al. (2004) Rationale and study design of the CardioGene Study: genomics of in-stent restenosis. Pharmacogenomics 5: 952–1004
Ganesh SK and Skelding KA et al., Genome-wide association study of in-stent restenosis identifies multiple candidate susceptibility loci. Presented at the AHA Scientific Sessions: 2006 November 12–15, Chicago
Faxon DP et al. (1984) Role of percutaneous transluminal coronary angioplasty in the treatment of unstable angina. Report from the National Heart, Lung, and Blood Institute Percutaneous Transluminal Coronary Angioplasty and Coronary Artery Surgery Study Registries. Am J Cardiol 53: 131C–135C
Lievers KJ et al. (2006) Association of a 31 bp VNTR in the CBS gene with postload homocysteine concentrations in the Framingham Offspring Study. Eur J Hum Genet 14: 1125–1129
Wang TH et al. (2005) Aspirin and clopidogrel resistance: an emerging clinical entity. Eur Heart J 27: 647–654
Shivakumar L and Armitage JO (2006) Bcl-2 gene expression as a predictor of outcome in diffuse large B-cell lymphoma. Clin Lymphoma Myeloma 6: 455–457
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Skelding, K., Gerhard, G., Simari, R. et al. The effect of HapMap on cardiovascular research and clinical practice. Nat Rev Cardiol 4, 136–142 (2007). https://doi.org/10.1038/ncpcardio0830
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncpcardio0830
This article is cited by
-
Mechanisms of Disease: the genetic basis of coronary heart disease
Nature Clinical Practice Cardiovascular Medicine (2007)