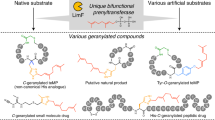
Abstract
This study highlights the biochemical and structural characterization of the L-tryptophan C6 C-prenyltransferase (C-PT) PriB from Streptomyces sp. RM-5-8. PriB was found to be uniquely permissive to a diverse array of prenyl donors and acceptors including daptomycin. Two additional PTs also produced novel prenylated daptomycins with improved antibacterial activities over the parent drug.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Walsh, C.T. Nat. Chem. Biol. 11, 620–624 (2015).
Tibrewal, N. & Tang, Y. Annu. Rev. Chem. Biomol. Eng. 5, 347–366 (2014).
Gantt, R.W., Peltier-Pain, P. & Thorson, J.S. Nat. Prod. Rep. 28, 1811–1853 (2011).
Elshahawi, S.I. et al. Proc. Natl. Acad. Sci. USA 110, E295–E304 (2013).
Ling, L.L. et al. Nature 517, 455–459 (2015).
Donia, M.S. & Fischbach, M.A. Science 349, 1254766 (2015).
Carr, G. et al. Org. Lett. 14, 2822–2825 (2012).
Wang, X. et al. Org. Lett. 17, 2796–2799 (2015).
Fan, A., Winkelblech, J. & Li, S.-M. Appl. Microbiol. Biotechnol. 99, 7399–7415 (2015).
Rudolf, J.D., Wang, H. & Poulter, C.D. J. Am. Chem. Soc. 135, 1895–1902 (2013).
Takahashi, S. et al. J. Bacteriol. 192, 2839–2851 (2010).
Winkelblech, J. & Li, S.M. ChemBioChem 15, 1030–1039 (2014).
Rudolf, J.D. & Poulter, C.D. ACS Chem. Biol. 8, 2707–2714 (2013).
Mori, T. et al. Nat. Commun. 7, 10849 (2016).
Subramanian, T., Liu, S., Troutman, J.M., Andres, D.A. & Spielmann, H.P. ChemBioChem 9, 2872–2882 (2008).
Eisenstein, B.I., Oleson, F.B. Jr. & Baltz, R.H. Clin. Infect. Dis. 50 (Suppl. 1): S10–S15 (2010).
Unsöld, I.A. & Li, S.-M. Microbiology 151, 1499–1505 (2005).
Mahmoodi, N. & Tanner, M.E. ChemBioChem 14, 2029–2037 (2013).
Yin, N. et al. J. Med. Chem. 58, 5137–5142 (2015).
Kuzuyama, T., Noel, J.P. & Richard, S.B. Nature 435, 983–987 (2005).
Bonitz, T., Alva, V., Saleh, O., Lupas, A.N. & Heide, L. PLoS One 6, e27336 (2011).
Tanner, M.E. Nat. Prod. Rep. 32, 88–101 (2015).
Williams, G.J., Zhang, C. & Thorson, J.S. Nat. Chem. Biol. 3, 657–662 (2007).
Nobeli, I., Favia, A.D. & Thornton, J.M. Nat. Biotechnol. 27, 157–167 (2009).
Feng, Y. et al. J. Am. Chem. Soc. 137, 10160–10163 (2015).
Chehade, K.A.H. et al. J. Am. Chem. Soc. 124, 8206–8219 (2002).
Subramanian, T., Wang, Z., Troutman, J.M., Andres, D.A. & Spielmann, H.P. Org. Lett. 7, 2109–2112 (2005).
Kearse, M. et al. Bioinformatics 28, 1647–1649 (2012).
Debono, M. et al. J. Antibiot. (Tokyo) 41, 1093–1105 (1988).
Shaaban, K.A. et al. J. Nat. Prod. 78, 1723–1729 (2015).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, Number 4. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Chen, V.B. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
This work was supported by NIH grants R37 AI52188 and R01 CA203257 (J.S.T.), U01 GM098248 (G.N.P.) and NCATS (UL1TR001998). Daptomycin (Cubicin) was generously provided by Merck. We are grateful to J. Rohr, S. Van Lanen and J. Chappell (College of Pharmacy, University of Kentucky) for helpful discussion and facilitating access to shared equipment and/or reagents. We thank the University of Kentucky Mass Spectrometry Facility for the HR–ESI–MS support. This research also used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science user facility operated by Argonne National Laboratory (DE-AC02-06CH11357). Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly and Company.
Author information
Authors and Affiliations
Contributions
S.I.E. and H.C. contributed to the experimental design and execution and manuscript preparation; K.A.S. and L.V.P. contributed to experimental design and execution; T.S. and H.P.S. contributed experimental reagents and consultation; M.L.F. contributed to experimental design and execution and provided key consultation; G.N.P. and J.S.T. contributed to the experimental design, project oversight and manuscript preparation; S.S. contributed to the experimental design and execution, project oversight and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
J.S.T. is a cofounder of Centrose (Madison, Wisonsin, USA).
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–28 and Supplementary Tables 1–11. (PDF 2642 kb)
Rights and permissions
About this article
Cite this article
Elshahawi, S., Cao, H., Shaaban, K. et al. Structure and specificity of a permissive bacterial C-prenyltransferase. Nat Chem Biol 13, 366–368 (2017). https://doi.org/10.1038/nchembio.2285
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2285
This article is cited by
-
Enzymatic β-elimination in natural product O- and C-glycoside deglycosylation
Nature Communications (2023)
-
Chemoenzymatic approaches for exploring structure–activity relationship studies of bioactive natural products
Nature Synthesis (2023)
-
Enzymatic formation of a prenyl β-carboline by a fungal indole prenyltransferase
Journal of Natural Medicines (2022)
-
Broadening the scope of biocatalytic C–C bond formation
Nature Reviews Chemistry (2020)
-
Enzymatic studies on aromatic prenyltransferases
Journal of Natural Medicines (2020)