Abstract
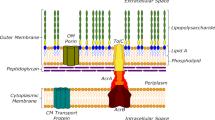
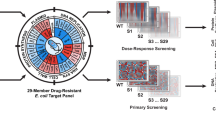
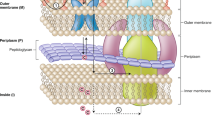
We developed a competition-based screening strategy to identify compounds that invert the selective advantage of antibiotic resistance. Using our assay, we screened over 19,000 compounds for the ability to select against the TetA tetracycline-resistance efflux pump in Escherichia coli and identified two hits, β-thujaplicin and disulfiram. Treating a tetracycline-resistant population with β-thujaplicin selects for loss of the resistance gene, enabling an effective second-phase treatment with doxycycline.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Referenced accessions
NCBI Reference Sequence
References
World Health Organization. Antimicrobial Resistance: Global Report on Surveillance (World Health Organization, 2014).
Andersson, D.I. & Hughes, D. Nat. Rev. Microbiol. 8, 260–271 (2010).
Lenski, R.E., Simpson, S.C. & Nguyen, T.T. J. Bacteriol. 176, 3140–3147 (1994).
Baym, M., Stone, L.K. & Kishony, R. Science 351, aad3292 (2016).
Chait, R., Craney, A. & Kishony, R. Nature 446, 668–671 (2007).
Palmer, A.C., Angelino, E. & Kishony, R. Nat. Chem. Biol. 6, 105–107 (2010).
Chait, R., Palmer, A.C., Yelin, I. & Kishony, R. Nat. Commun. 7, 10333 (2016).
Imamovic, L. & Sommer, M.O.A. Sci. Transl. Med. 5, 204ra132 (2013).
Szybalski, W. & Bryson, V. J. Bacteriol. 64, 489–499 (1952).
Lázár, V. et al. Mol. Syst. Biol. 9, 700 (2013).
Hiramatsu, K. et al. Int. J. Antimicrob. Agents 39, 478–485 (2012).
Chao, L. Nature 271, 385–386 (1978).
Lukens, A.K. et al. Proc. Natl. Acad. Sci. USA 111, 799–804 (2014).
Gonzales, P.R. et al. Nat. Chem. Biol. 11, 855–861 (2015).
Kim, S., Lieberman, T.D. & Kishony, R. Proc. Natl. Acad. Sci. USA 111, 14494–14499 (2014).
Bochner, B.R., Huang, H.C., Schieven, G.L. & Ames, B.N. J. Bacteriol. 143, 926–933 (1980).
Merlin, T.L., Davis, G.E., Anderson, W.L., Moyzis, R.K. & Griffith, J.K. Antimicrob. Agents Chemother. 33, 1549–1552 (1989).
Stone, G.W. et al. Antimicrob. Agents Chemother. 48, 477–483 (2004).
Wright, G.D. Chem. Commun. 47, 4055–4061 (2011).
Chait, R., Shrestha, S., Shah, A.K., Michel, J.-B. & Kishony, R. PLoS One 5, e15179 (2010).
Chopra, I. & Roberts, M. Microbiol. Mol. Biol. Rev. 65, 232–260 (2001).
Wang, H. & Ng, T.B. Life Sci. 65, 849–856 (1999).
Ejim, L. et al. Nat. Chem. Biol. 7, 348–350 (2011).
Anderson, A.B. & Gripenberg, J. Acta Chem. Scand. 2, 644–650 (1948).
Phillips, M., Malloy, G., Nedunchezian, D., Lukrec, A. & Howard, R.G. Antimicrob. Agents Chemother. 35, 785–787 (1991).
Alekshun, M.N. & Levy, S.B. Cell 128, 1037–1050 (2007).
Katayama, Y., Ito, T. & Hiramatsu, K. Antimicrob. Agents Chemother. 44, 1549–1555 (2000).
Meyer, B.J., Maurer, R. & Ptashne, M. J. Mol. Biol. 139, 163–194 (1980).
Lutz, R. & Bujard, H. Nucleic Acids Res. 25, 1203–1210 (1997).
Baym, M. et al. PLoS One 10, e0128036 (2015).
Martin, M. EMBnet.Journal 17, 10–11 (2011).
Langmead, B. & Salzberg, S.L. Nat. Meth. 9, 357–359 (2012).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Albers, C.A. et al. Genome Res. 21, 961–973 (2011).
Keane, T.M., Wong, K. & Adams, D.J. Bioinformatics 29, 389–390 (2013).
Acknowledgements
We thank D. Rudner, R. Mazitschek, and R. Moellering for helpful insights; J. Horn and J. Marchionna for custom screening plate manufacture and technical advice; D. Flood and S. Rudnicki for technical support in the primary screen; and J. Wang and J. Moore for technical support in the flow cytometry assay. All primary screening was performed at Harvard Medical School ICCB-L Screening Facility. This work was supported in part by National Institute of Allergy and Infectious Diseases grant U54 AI057159, US National Institutes of Health grants R01 GM081617 (to R.K.) and GM086258 (to J.C.), European Research Council FP7 ERC grant 281891 (to R.K.) and a National Science Foundation Graduate Fellowship (to L.K.S.).
Author information
Authors and Affiliations
Contributions
L.K.S., J.C., and R.K. designed research; L.K.S. performed experiments and analyzed data; M.B. and R.C. built the imaging setup and M.B. developed the automation; L.K.S. and M.B. performed genomic sequencing; T.D.L. analyzed genomic sequencing data; R.C. contributed the initial plate and assay design; L.K.S. and R.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–14 and Supplementary Tables 1–6. (PDF 24325 kb)
Rights and permissions
About this article
Cite this article
Stone, L., Baym, M., Lieberman, T. et al. Compounds that select against the tetracycline-resistance efflux pump. Nat Chem Biol 12, 902–904 (2016). https://doi.org/10.1038/nchembio.2176
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2176
This article is cited by
-
Collateral responses to classical cytotoxic chemotherapies are heterogeneous and sensitivities are sparse
Scientific Reports (2022)
-
Antibiotic combinations reduce Staphylococcus aureus clearance
Nature (2022)
-
Rapid expansion and extinction of antibiotic resistance mutations during treatment of acute bacterial respiratory infections
Nature Communications (2022)
-
Ubiquitous selection for mecA in community-associated MRSA across diverse chemical environments
Nature Communications (2020)
-
Bacterial variability in the mammalian gut captured by a single-cell synthetic oscillator
Nature Communications (2019)