Abstract
Transforming growth factor-β (TGF-β) induces the expression of Disabled-2 (Dab2), an endocytic adaptor and tumour suppressor, concomitant with the induction of an epithelial–mesenchymal transition (EMT) in mammary epithelial cells. Here we show that following TGF-β-mediated EMT, sustained TGF-β treatment leads to proteolytic degradation of Dab2 by cathepsin B (CTSB), loss of the mesenchymal phenotype and induction of autophagy. CTSB inhibition or expression of a CTSB-resistant Dab2 mutant maintains Dab2 expression and shifts long-term TGF-β-treated cells from autophagy to apoptosis. We further show that Dab2 interacts with Beclin-1 to promote casein-kinase-2-mediated phosphorylation of Beclin-1, preventing Beclin-1–Vps34 interaction and subsequent autophagosome assembly. Thus, CTSB-mediated degradation of Dab2 allows Beclin-1–Vps34 induction of autophagy, whereas sustained Dab2 expression prevents autophagy and promotes apoptosis by stabilizing the pro-apoptotic Bim protein. In vivo studies suggest that Dab2-mediated regulation of autophagy modulates chemotherapeutic resistance and tumour metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Massague, J. TGFβ in cancer. Cell 134, 215–230 (2008).
Fischer, K. R. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015).
Zheng, X. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Suzuki, H. I., Kiyono, K. & Miyazono, K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy 6, 645–647 (2010).
Avalos, Y. et al. Tumor suppression and promotion by autophagy. BioMed Res. Int. 2014, 603980 (2014).
Gozuacik, D. & Kimchi, A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23, 2891–2906 (2004).
Degenhardt, K. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 (2006).
Hannigan, A. et al. Epigenetic downregulation of human disabled homolog 2 switches TGF-β from a tumor suppressor to a tumor promoter. J. Clin. Invest. 120, 2842–2857 (2010).
Chaudhury, A. et al. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 (2010).
O’Connor, L. et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 (1998).
Edinger, A. L. & Thompson, C. B. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 16, 663–669 (2004).
Xu, X. X., Yang, W., Jackowski, S. & Rock, C. O. Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J. Biol. Chem. 270, 14184–14191 (1995).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Luciano, F. et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22, 6785–6793 (2003).
Jiang, Y., He, X. & Howe, P. H. Disabled-2 (Dab2) inhibits Wnt/β-catenin signalling by binding LRP6 and promoting its internalization through clathrin. EMBO J. 31, 2336–2349 (2012).
Pinna, L. A. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115, 3873–3878 (2002).
Itakura, E., Kishi, C., Inoue, K. & Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 (2008).
Matsunaga, K. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 (2009).
Simonsen, A. & Tooze, S. A. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186, 773–782 (2009).
Kim, J. et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 (2013).
Dote, H. et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin. Cancer Res. 10, 2082–2089 (2004).
Podgorski, I. & Sloane, B. F. Cathepsin B and its role(s) in cancer progression. Biochem. Soc. Symp. 70, 263–276 (2003).
Sinha, A. A., Jamuar, M. P., Wilson, M. J., Rozhin, J. & Sloane, B. F. Plasma membrane association of cathepsin B in human prostate cancer: biochemical and immunogold electron microscopic analysis. Prostate 49, 172–184 (2001).
Sloane, B. F. Cathepsin B and cystatins: evidence for a role in cancer progression. Semin. Cancer Biol. 1, 137–152 (1990).
Werle, B., Jülke, B., Lah, T., Spiess, E. & Ebert, W. Cathepsin B fraction active at physiological pH of 7.5 is of prognostic significance in squamous cell carcinoma of human lung. Br. J. Cancer 75, 1137–1143 (1997).
Linebaugh, B. E., Sameni, M., Day, N. A., Sloane, B. F. & Keppler, D. Exocytosis of active cathepsin B enzyme activity at pH 7.0, inhibition and molecular mass. Eur. J. Biochem. 264, 100–109 (1999).
Mort, J. S., Recklies, A. D. & Poole, A. R. Extracellular presence of the lysosomal proteinase cathepsin B in rheumatoid synovium and its activity at neutral pH. Arthritis Rheum. 27, 509–515 (1984).
Acknowledgements
We thank G. Hussey, B. Howley, T. Smith and other members of our laboratory for helpful comments and critical insights. We are very grateful to B. Levine (Center for Autophagy Research, University of Texas Southwestern Medical Center, USA) for providing us with Beclin-1 constructs. The work was supported in part by the Flow Cytometry & Cell Sorting, Molecular Imaging, Small Animal Imaging, and the Gene Targeting and Knockout Shared Resources, Hollings Cancer Center, Medical University of South Carolina (P30CA138313). Our study used the services of the MUSC Center for Oral Health Research (COHR), which is partially supported by the National Institute of General Medicine grant P30GM103331. This work was also supported by grants CA555536 and CA154664 from the National Cancer Institute to P.H.H.
Author information
Authors and Affiliations
Contributions
Y.J. and P.H.H. conceived of the project and designed the experiments. Y.J., A.N.W., N.S. and S.N. performed experiments. Y.J. and P.H.H. analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Identification of the CTSB cleavage site in Dab2 and the TGFβ signaling pathway for CTSB regulation.
(a) RT-PCR analysis of p96 Dab2, p67 Dab2, cathepsin B and β-actin mRNA levels in NMuMG cells. (b) Eph4 Ras cells and HMLE cells were treated with TGFβ for 7 days and immunoblots were performed to detect the expression of Dab2 and cathepsin B. Meanwhile, LC3B level was compared by immunoblotting between 0 day and 7 days treatment in the presence and absence of chloroquine. (c) Inhibitor screen to identify the protease responsible for Dab2 cleavage. Each inhibitor was added to cells at the concentrations indicated for 6 h prior to the preparation of whole cell lysates and immunoblot analysis. (d) Schematic of wild-type, full length Dab2, Dab2 mutants and amino acid sequence of peptides 486-510. (e) In vitro cleavage analysis of [35S]-methionine-labeled p96 Dab2 and Dab2 mutants. (f) [35S]-methionine-labeled wild-type and Dab2 mutants shown in (d) were incubated for 1 h at 37 °C with extracts from cells treated with TGFβ for 7 days prior to the preparation of WCLs and autoradiographic analysis of p96 Dab2 cleavage. (g) Immunoblot analysis of total and phosphorylated Smad2 and Smad3 following a 7-day TGFβ treatment. (h) Immunoblot analysis of total Smad2 and Smad3 following shRNA-mediated silencing. Hsp90 served as a loading control. (i) RT-PCR analysis of cathepsin B and β-actin mRNA levels in control and Smad2/3 double-knockdown NMuMG cells (top panel), and immunoblot analysis of cathepsin B, N-cadherin, and vimentin (lower panel) following a 7-day TGFβ treatment. (j) Nuclear, cytosolic and membrane fractions isolated from NMuMG cells treated with TGFβ were subjected to immunoblot analysis to detect the expression of cathepsin B, sodium potassium ATPase alpha1 (membrane fraction marker), histone H3 (nuclear fraction marker) and α-tubulin (cytosolic fraction marker). (k) CTSB activity assay in vitro using extracts from control and Smad2/3 double knockdown NMuMG cells treated with TGFβ for the times indicated. Data was shown as mean ± s.d., n = 3 independent experiments. The experiments were repeated three times and similar results were observed. Unprocessed blots are available in Supplementary Figure 9.
Supplementary Figure 2 Chronic TGFβ treatment results in autophagy and the reversal of EMT.
(a) NMuMG and NMuMG CTSB/OE cells were treated with TGF-β for 5 and 7 days. Control and 25 μm chloroquine solutions were applied to cell culture media 3 h before collecting the cell lysate of each time point. Cell lysates were subjected to immunoblot to detect LC3B expression with a α-LC3B antibody. LC3BII levels were quantified from n = 3 independent experiments, shown as mean ± s.d. (b) The GFP-LC3B puncta from top panel of figure 2b was quantified from n = 3 independent experiments, shown as mean ± s.d. (c) The expression of autophagy markers (LC3BII and p62) and apoptosis markers (Bim and cleaved caspase-3) of figure 3b & c was quantified using a STORM scanner and ImageQuant software from Molecular Dynamics (Fairfield, CT, USA). Levels are expressed as relative intensities. The data is presented as the mean ± s.d., n = 3 independent experiments. (d) Cell death rates were quantified from figure 3d experiments. The data is presented as the mean ± s.d. n = 3 independent experiments. (e) Whole cell lysates from Dab2 and CTSB-modulated NMuMG cells treated with TGFβ for the times indicated were subjected to immunoblot analysis to determine the expression of mesenchymal markers and cell invasion markers using their corresponding antibodies. All experiments were repeated three times and similar results were observed. Unprocessed blots are available in Supplementary Figure 9.
Supplementary Figure 3 Dab2 blocks ERK/Bim interaction and attenuates ERK-mediated Bim phosphorylation and degradation.
(a) Co-immunoprecipitation analysis using a α-ERK1/2 or α-phospho-ERK1/2 antibody of whole cell lysates from NMuMG/OE ± LVLDab2 cells treated with TGFβ ± PYR-41 for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of p-ERK1/2 and Bim. (b) Immunoblotting analysis was applied to detect Bim and p-Bim expression from whole cell lysates of NMuMG/OE ± LVLDab2 cells treated with TGFβ for the times indicated in the presence of 20 μM PYR-41 (applied 4 h before cell lysates were collected). (c,d) Immunofluorescence analysis was performed to detect the co-localization between ERK or p-ERK and Bim before and after NMuMG/OE ± LVLDab2 cells were treated for 7 days with TGFβ. Scale bar, 10 μm. The experiments were repeated three times and similar results were observed. Unprocessed blots are available in Supplementary Figure 9.
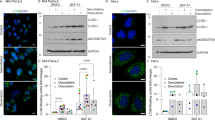
Supplementary Figure 4 Characterization of Beclin-1/Dab2 interactions and followed PI3K induction.
(a) Mapping the interaction domain between Dab2 and Beclin-1 in NMuMG CTSB/KD cells. Schematic diagram of domains of Beclin-1 and schematic representatives of Flag-tagged wild type Beclin-1 and mutant Beclin-1 constructs employed in co-immunoprecipitation assay (left). Co-immunoprecipitation assays of transfected Flag-tagged wild type Beclin-1 and Beclin-1 mutants with Dab2 (right). (b) Immunofluorescence analysis of CK2 and Beclin-1 in NMuMG CTSB/OE cells and NMuMG CTSB/OE + LVLDab2 cells. CK2 is shown as green, and Beclin-1 is shown as red. Colocalization of CK2 and Beclin-1 is depicted as yellow in the panels labeled ‘merge’. DAPI was used to stain nuclei. Scale bars, 10 μm. (c) Inhibition of CK2 attenuates Dab2’s inhibitory effect on Beclin-1/Vps34 interaction. Co-immunoprecipitation analysis using a α-Beclin-1 antibody of whole cell lysates from CTSB/OE and CTSB/OE + LVLDab2 cells treated with TGFβ for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of Vps34, Dab2, CK2, and Beclin-1. Immunoblots (IB) depict expression levels of Vps34, Dab2, and Beclin-1 in whole cell lysates. The CK2 inhibitor apigenin (10 and 20 μM) was added 6 h prior to the preparation of whole cell lysates. (d) Analysis of LC3BII turnover in the absence and presence of chloroquine. NMuMG/LVLDab2 cells stably transfected with Beclin-1 S337A/S341A mutant were subjected to TGFβ treatment as indicated. Control and 25 μM chloroquine solutions were applied to cell culture media 3 h before collecting the cell lysate of each time point. Cell lysates were subjected to immunoblot to detect LC3B expression with a α-LC3B antibody (left panel), and the quantification of LC3BII (Fig. 6g) is shown in the right panel as mean ± s.d., n = 4 independent experiments. (e) PI3P levels before and after TGFβ treatment in NMuMG cells (left) and NMuMG/LVLDab2 cells (right) with Beclin-1 knockdown and ectopic expression of WT Beclin-1 and mutant Beclin-1. Data was shown as mean ± s.d., n = 4 independent experiments. The experiments were repeated four times and similar results were observed. Unprocessed blots are available in Supplementary Figure 9.
Supplementary Figure 5 Transmission electron microscopy (TEM) analysis.
Transmission electron microscopy analysis (TEM) was performed to detect autophagic and apoptotic structures in (a) MTT assay depicting the dose dependent effects of doxorubicin (DOXO) on cell survival in NMuMG, BT20, MDA-MB468 cells, and their modified derivatives. Results are shown as means −/+ s.d., n = 3 independent experiments. (b) NMuMG control cells, CTSB/OE cells and CTSB/OE + LVLDab2 cells, in (c) BT-20 control, Dab2 KD and muBeclin-1 overexpressing cells and in (d) MDA-MB-468 control, LVLDab2 and LVLDab2 + muBeclin-1 overexpressing cells, before and after doxorubicin (DOXO) treatment. Scale bars, 2 μm. Frame sections highlight autophagosomes. (e) NMuMG, BT20, MDA-MB-468 cells and their transfected derivatives were subjected to DOXO (2 μg/ml) for 24 h treatment. 25 μM chloroquine was applied to cell culture media 1 h before cell lysates were collected. Cell lysates were subjected to immunoblotting analysis to detect LC3B expression with α-LC3B antibody. (f) NMuMG, BT20, MDA-MB-468 cells and their transfected derivatives were treated with DOXO (2 μg/ml) for 12 h and 24 h. Cell lysates were subjected to immunoblotting analysis to detect Bim expression with α-Bim antibody. Hsp90 served as loading control. The experiments were repeated three times and similar results were observed. Unprocessed blots are available in Supplementary Figure 9.
Supplementary Figure 6 Dab2 attenuates doxorubicin-induced Beclin-1/Vps34 interactions.
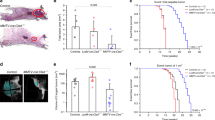
(a) Immunoprecipitation analysis was performed with a α-Beclin-1 antibody from whole cell lysates of NMuMG/OE ± LVLDab2 cells, BT-20 control cells and Dab2 KD cells, and MDA-MB-468 control ± LVLDab2 cells to detect Beclin-1/Vps34 interactions during DOXO treatment. Immunoprecipitated complexes were immunoblotted to detect the expression of Dab2 and Vps34. (b) Immunoblotting analysis of LC3B and Bim in tissue lysates from tumors of Figure 8a with α-LC3B and α-Bim antibodies. Chloroquine was injected 12 h before the extraction of the tumors (i.v., 20 mg/kg) (c) Quantification of the LC3B and Bim expression in panel b. Data represents the means −/+ s.d., n = 3 independent experiments. Unprocessed blots are available in Supplementary Figure 9.
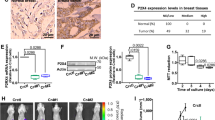
Supplementary Figure 7 Immunohistochemical analysis of primary tumors from BT20, MDA-MB-468 and their transfected derivatives.
Representatives of immunohistochemical staining of Hematoxylin and eosin (H&E), Ki-67 TUNEL, p62, Dab2, Cathepsin B and Flag-tagged muBeclin-1 in primary tumors from BT-20 and MDA-MB-468 and their derivatives before and after DOXO treatment to the mice. Scale bar, 25 μm. All experiments were repeated three times and similar results were observed.
Supplementary Figure 8 The Immunhistochemical analysis of metastases and the working model.
(a) Immunohistochemical analysis of metastases. Representative immunohistochemical staining of Hematoxylin and eosin (H&E), Ki-67 TUNEL, p62, Dab2, Cathepsin B and Flag-tagged muBeclin-1 of metastatic lesions developed from BT-20 Dab2 KD cells or BT-20 muBeclin-1 overexpressing cells (liver) and from MDA-MB-468 control cells or muBeclin-1/LVLDab2 overexpressing cells (lung). Scale bar, 25 μm. (b) Model depicting Dab2’s role in mediating autophagy or apoptosis in chronic TGFβ-treated cells. Dab2 inhibits TGFβ-mediated autophagy by attenuating Beclin-1/Vps34 interactions. Dab2 prevents ERK-mediated Bim phosphorylation and degradation and stabilizes TGFβ-induced Bim expression to promote apoptosis. The experiments were repeated three times and similar results were observed.
Supplementary information
Supplementary Information
Supplementary Information (PDF 10805 kb)
Supplementary Table 1
Supplementary Information (XLSX 32 kb)
Rights and permissions
About this article
Cite this article
Jiang, Y., Woosley, A., Sivalingam, N. et al. Cathepsin-B-mediated cleavage of Disabled-2 regulates TGF-β-induced autophagy. Nat Cell Biol 18, 851–863 (2016). https://doi.org/10.1038/ncb3388
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3388
This article is cited by
-
Cathepsin B in programmed cell death machinery: mechanisms of execution and regulatory pathways
Cell Death & Disease (2023)
-
Autophagy deficiency promotes lung metastasis of prostate cancer via stabilization of TWIST1
Clinical and Translational Oncology (2022)
-
TFEB-driven autophagy potentiates TGF-β induced migration in pancreatic cancer cells
Journal of Experimental & Clinical Cancer Research (2019)
-
A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens
Nature Biomedical Engineering (2019)
-
Dpp regulates autophagy-dependent midgut removal and signals to block ecdysone production
Cell Death & Differentiation (2019)