Abstract
Epigenetic consequences of exposure to psychostimulants are substantial but the relationship of these changes to compulsive drug taking and abstinence is not clear. Here, we used a paradigm that helped to segregate rats that reduce or stop their methamphetamine (METH) intake (nonaddicted) from those that continue to take the drug compulsively (addicted) in the presence of footshocks. We used that model to investigate potential alterations in global DNA hydroxymethylation in the nucleus accumbens (NAc) because neuroplastic changes in the NAc may participate in the development and maintenance of drug-taking behaviors. We found that METH-addicted rats did indeed show differential DNA hydroxymethylation in comparison with both control and nonaddicted rats. Nonaddicted rats also showed differences from control rats. Differential DNA hydroxymethylation observed in addicted rats occurred mostly at intergenic sites located on long and short interspersed elements. Interestingly, differentially hydroxymethylated regions in genes encoding voltage (Kv1.1, Kv1.2, Kvb1 and Kv2.2)- and calcium (Kcnma1, Kcnn1 and Kcnn2)-gated potassium channels observed in the NAc of nonaddicted rats were accompanied by increased mRNA levels of these potassium channels when compared with mRNA expression in METH-addicted rats. These observations indicate that changes in differentially hydroxymethylated regions and increased expression of specific potassium channels in the NAc may promote abstinence from drug-taking behaviors. Thus, activation of specific subclasses of voltage- and/or calcium-gated potassium channels may provide an important approach to the beneficial treatment for METH addiction.
Similar content being viewed by others
Introduction
Drug self-administration (SA) offers an important model to investigate transcriptional and epigenetic programs that may drive the transition to addicted states after initial drug exposure.1, 2, 3 Behavioral phenomena observed during drug SA are regulated by a diversity of interconnected but distinct brain regions that include the nucleus accumbens (NAc) and dorsal striatum, among others.4, 5, 6 The NAc is an important link in the circuit of addiction because it connects to brain regions that subsume positive memory7 and decision making,8 two cognitive processes that regulate drug-taking behaviors in rodents and humans.9, 10 Indeed, pharmacological manipulations that affect some of these structures have been shown to increase, attenuate or stop animals from self-administering various drugs of abuse.11, 12 Among these brain regions, the NAc appears to play critical roles in the initiation and maintenance of drug self-administration.13 Importantly, dopaminergic neurons from the ventral tegmental area14 and glutamatergic neurons from the prefrontal cortex (PFC)15 send projections to the NAc and can regulate plastic mechanisms that form the substrates of addiction.1, 6 These neuroplastic changes appear to occur via alterations of transcripts and protein levels in intrinsic NAc neurons.16, 17, 18, 19
Similar to other psychostimulants, methamphetamine (METH) addiction is highly prevalent throughout the world and is associated with a diversity of neuropsychiatric signs and symptoms.20 Some of the long-term effects of this drug may be the results of molecular events that occur in the rodent striatum and NAc after either contingent or noncontingent administration of the drug.20, 21, 22, 23 In addition to changes in the expression of immediate early genes,4, 22 METH can also alter the expression of several neuropeptides including prodynorphin in the METH SA model.23 Nevertheless, experimental approaches that use all rats that self-administer a drug to measure biochemical or physiological consequences of drugs do not strictly model human conditions24, 25, 26 because not all humans who use rewarding drugs become addicted to them. Importantly, criteria for the addiction diagnosis include the persistence of drug taking in the presence of adverse consequences (American Psychiatric Association (APA) and Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM V)) by definition. It is also clear that a substantial number of individuals who have experimented with drugs can self-impose abstinence because of psychosocial complications associated with drug use.27
In order to investigate epigenetic dysfunctions that may subsume METH addiction, we used a model of compulsive drug taking that was recently shown to produce a subgroup of rats that continue to lever press compulsively for cocaine despite adverse consequences.28, 29, 30 Here, we show that there also exists a subpopulation of rats that continue to lever press for METH even in the presence of footshocks, whereas other rats became progressively more abstinent with increasing shock intensity. Moreover, we discovered that rats that differ phenotypically also exhibited substantial differences in DNA hydroxymethylation in peaks located near or within genes that are relevant to synaptic plasticity and cognition.
Materials and methods
Subjects
Male Sprague–Dawley rats, 14–16 weeks old (Charles River, Raleigh, NC, USA), weighing 350–400 g before surgery, were used in our experiments. Rats were housed in the animal facility under a reversed 12:12 h light/dark cycle with food and water available ad libitum except during the first 1–2 days of self-administration training when regular home cage food was limited to 20 g per day. All procedures followed the guidelines outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (eighth edition; https://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-use-of-laboratory-animals.pdf) and were approved by the local NIDA (National Institute on Drug Abuse) animal care committee.
Intravenous surgery
Surgery for the implantation of intravenous catheter was done essentially as described before.26, 31 Rats were anesthetized with ketamine (50 mg kg−1, intraperitoneal) and xylazine (5 mg kg−1, intraperitoneal) and inserted with silastic catheters into the jugular vein. The catheters were attached to a modified 22-gauge cannula that was mounted to the rats’ skulls with dental cement. We injected buprenorphine (0.1 mg kg−1, subcutaneous) after surgery to relieve pain and allowed the rats to recover for 7 days before METH SA training. During the recovery, training and punishment phases, we flushed the catheters every 24–48 h with gentamicin (Henry Schein, Melville, NY, USA; 5 mg ml−1) and sterile saline.
Apparatus
Rats were trained in SA chambers located inside sound-attenuating cabinets and controlled by a Med Associates System (Med Associates, St Albans, VT, USA). Each chamber was equipped with two levers located 8.5 cm above the grid floor and the grid floors were connected to electric shock generators. Presses on the retractable active lever activated the infusion pump. Presses on the inactive lever had no reinforced consequences. The catheters of rats were connected to an intravenous line (Plastics One, Roanoke, VA, USA) attached to a liquid swivel (Instech, Plymouth, PA, USA) via polyethylene-50 tubing that was protected by a metal spring.
Training phase
The training procedure for METH SA was similar to that used in previous studies.26 On the first day of training, the rats were brought to the SA room where they were chronically housed in SA chambers. Rats were randomly assigned to either METH SA or control groups. Rats (n=24) were trained to self-administer dl-METH HCl (NIDA) during three 3-h sessions per day (the sessions were separated by 30 min) over 20 days under a fixed-ratio 1 with a 20-s timeout reinforcement schedule (Figure 1a). Lever presses were accompanied by a 5-s compound tone–light cue. We trained the rats in cycles of 2 days of drug SA and 1 day off in order to control for weight loss, a common side effect of METH in laboratory animals.32 METH was dissolved in sterile saline and the rats self-administered the drug at a dose of 0.1 mg kg−1 per infusion over 3.5 s (0.1 ml per infusion).26 Control rats (n=10) self-administered saline (0.1 ml per infusion). The number of infusions per 3 h session was limited to 35 in order to control for potential overdosing. At the end of each 3 h session, the housing light was turned off, and the active lever was retracted.
Prolonged methamphetamine (METH) self-administration and contingent footshock produce rats with nonaddicted and compulsive drug-taking phenotypes. (a) Timeline of the experiment. (b) All rats escalate their intake of METH during the training phase of the experiment. The inset shows that shock-resistant (SR) and shock-sensitive (SS) rats took similar amount of METH before the shock phase. (c) Footshocks suppress lever pressing in SS rats but not in SR compulsive METH takers. The inset illustrates the fact that SS rats took substantially less METH than SR rats during that phase. (d) Lever pressing on the last day of training and last day of footshocks. #P<0.05, ###P<0.001, in comparison with SR rats; ***P<0.001 in comparison with last day of training.
Footshock phase
During the shock phase, the rats continued to self-administer METH every day (9 h sessions) under the same fixed-ratio 1, 20-s timeout reinforcement schedule used during training. In addition, 50% of the reinforced active lever presses resulted in the concurrent delivery of a 0.5-s footshock through the grid floor.26 The initial footshock was set at 0.18 mA and was increased by 0.06 mA daily up to 0.30 mA. Thereafter, the rats received footshocks of 0.30 mA for 4 days followed by 0.36 mA for another 4 days. We chose to increase the shock progressively because adverse conditions in humans progress over time. Punished responses continued to produce the tone–light cue and 0.1 mg kg−1 per infusion of METH.
Tissue collection
We killed the rats at 2 h after the last shock session. Brains were removed from the skull and the NAc was collected to be used in hydroxymethylated DNA immunoprecipitation (hMeDIP) sequencing (hmC-Seq) and quantitative PCR analysis that are described in detail in the Supplementary Methods.
Analysis of hmC-Seq data
A combinatorial approach was used for high confidence peak calling. Statistically significant peaks of mapped reads were detected by using the model-based analysis of ChIP-Seq (MACS) program33 on each hmC-Seq file against the matching input file under the stringent condition that satisfies fold enrichment ⩾10 and the false discovery rate ⩽0.05%. Replicates were combined by retaining only those peaks that meet the majority rule condition (>50% of samples identify a peak) as described previously.34 To measure the reproducibility at the level of peak calling, irreproducible discovery rate (IDR) analysis35 was applied to the sets of peaks identified from replicates. Next, peaks were identified using PePr36 (peak-calling prioritization pipeline) that uses a negative binomial distribution to identify consistent or differential binding sites in ChIP-Seq experiments with biological replicates. Peaks called in both MACS2-IDR and PePr pipelines were considered as high confidence calls.
Peak annotations were performed using HOMER (hypergeometric optimization of motif enrichment).37 The annotation includes peaks in the transcription start site, transcription termination site, Exon (Coding), 5′-untranslated region exon, 3' untranslated region exon or intron. The locations outside these were defined as intergenic regions and include the nearest promoter, nearest gene, distance to transcription start site and motif occurrences in peaks. In addition to associating peaks with nearby genes, the annotation includes the Gene Ontology Analysis and the Genomic Feature association analysis that look for enrichment of various gene function and genomic annotations in the list of peaks/regions, respectively.
Pathway analysis
We used Ingenuity Pathway Analysis (IPA) software (Qiagen, Valencia, CA, USA) to identify molecular functions, canonical pathways and biological networks. We also used the PANTHER (protein annotation through evolutionary relationships) classification system to analyze genes that showed differential DNA hydroxymethylation (http://www.pantherdb.org/).38
Statistical analyses
We analyzed the behavioral data with the statistical program SPSS (IBM, NY, USA) and followed significant effects (P<0.05) with SPSS post hoc contrasts within the repeated measures analysis of variance module. For the training and footshock phases, the dependent variables were the number of METH infusions during the training days and the punishment sessions. The dependent variables for the extinction tests were total (nonreinforced) active lever presses and inactive lever presses.
Results
Training phase
Figure 1a shows the timeline for the drug SA experiments. All METH-trained rats significantly escalated their intake of the drug per session during the first 16 days and then maintained their intake for the remainder of the training session (Figure 1b). The repeated measures mixed analysis of variance for rewards earned included the between-subject factor group (control (CT), shock-sensitive (SS), shock-resistant (SR)) and the within-subject factor of day (training days 1–20). This analysis showed a significant effect of day × group (F(38, 608)=18.4, P<0.001). The significant interaction indicates that METH intake continued to increase for the first 16 training days for METH-trained rats, whereas saline intake decreased and stabilized after 7 days for control rats. There were no significant differences in reward acquisition during training between METH-trained SS and SR groups (P>0.05; Figure 1b).
Footshock phase
During the footshock phase, we increased shock intensity over several days (Figure 1c). The shocks led to the segregation of the METH-trained animals into two phenotypes. One group continued to compulsively press the lever for METH, whereas the other group progressively decreased their intake (Figure 1c). As the intensity of footshocks increased over days, METH-reinforced responding decreased in SS (nonaddicted) but not in SR (METH-addicted) group. The statistical analysis of METH infusions earned by SS and SR rats included the between-subject factor of group (SS, SR) and within-subject factor of shock day (shock days 1–10). There was a significant effect of shock day × group (F(9, 198)= 5. 09, P<0.001), indicating that addicted rats self-administered more METH infusions than nonaddicted rats, an effect that was most pronounced at the higher shock dose (0.36 mA) (Figure 1c). Statistical analysis showed no significant differences in the number of METH infusions between SS and SR groups at the end of SA training (Figure 1d). However, in the end of footshock phase, SS rats dramatically reduced the number of METH infusions in comparison with their preshock intake and in comparison with SR rats. In contrast, SR rats did not reduce the number of METH infusions versus their preshock level (Figure 1d).
DNA hydroxymethylation
At 2 h following the SA and footshock regimen, the animals were killed and genomic DNA was extracted from the NAc. Isolated DNA was immunoprecipitated using an antibody against hydroxymethylated cytosine. Immunoprecipitated DNA was made into sequencing libraries and subjected to next-generation sequencing. Reads generated from sequencing underwent quality control measures (see Supplementary Methods) and were aligned to the rat genome (UCSC (University of California, Santa Cruz) Genome Browser rn5). The MACS2 peak-calling program was used to convert loci with enriched reads as peaks. We considered peaks that satisfied fold enrichment ⩾10 and the false discovery rate ⩽0.05. In addition, we chose peaks that exist for more than three of the six animals per group. Using the criteria described above, comparisons were performed between SR and CT (SRvsCT), SS and CT (SSvsCT) and SR and SS (SRvsSS) groups. The hMeDIP Seq data set has been deposited and is freely available at http://www.ncbi.nlm.nih.gov/bioproject/310210.
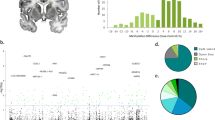
Figure 2 and Supplementary Table S1 show the results of the overall DNA hydroxymethylation experiments. We found significant differentially hydroxymethylated peaks between SRvsCT, SSvsCT and SRvsSS comparisons. There were 475 (427 hyper- and 48 hypo-hydroxymethylated) peaks that showed differential DNA hydroxymethylation regions in the SRvsCT comparison, 808 (797 hyper and 11 hypo) peaks in the SSvsCT comparison and 992 (598 hyper and 394 hypo) peaks in the SRvsSS comparison (Figure 3 and Supplementary Figures S1 and S2). The Venn diagrams also show the overlaps among the three comparisons for the number of total genes (Figure 2a) and intragenic genes (Figure 2b). Figure 2a shows that there were 211 genes with differentially hydroxymethylated peaks unique to the SRvsCT comparison, 479 genes restricted to the SSvsCT comparison and 547 genes in the SRvsSS comparison, indicating that METH-addicted and nonaddicted rats could also be segregated based on their genome-wide DNA hydroxymethylation profiles.
Whole genome profiling in methamphetamine (METH)-addicted (shock-resistant (SR)) and nonaddicted (shock-sensitive (SS)) rats. The Venn diagrams show the number of (a) all hydroxymethylated peaks and (b) only intragenic peaks in the different METH and footshock-induced phenotypes. (c) Distribution of hydroxymethylated peaks in the three pairwise comparisons.
Functional annotation of differentially hydroxymethylated peaks between METH-addicted (shock-resistant (SR)) and nonaddicted (shock-sensitive (SS)) rats. The data sets were grouped according to (a) hyper-hydroxymethylated and (b) hypo-hydroxymethylated peaks in the SRvsSS comparison. Gene families were grouped based on their molecular functions, biological processes and protein classes according to the PANTHER analysis. Differences in hydroxymethylated peaks were considered significant as described in the text. The genes with differentially hydroxymethylated peaks are listed in Supplementary Table S1.
Figure 2c shows the distribution of differentially hydroxymethylated peaks in the three pairwise comparisons. The majority of peaks were found in intergenic sites and in repeat elements. There were very few changes located in transcription start sites or in exons. Interestingly, many changes occurred in long interspersed elements,39 with the smaller number of changes occurring in the SSvsCT comparison (27.9% against 56.2% and 55.6% for SRvsCT and SRvsSS, respectively) (Figure 2c). Changes in DNA hydroxymethylation also occurred in short interspersed elements, with peak counts being similar across the three comparisons.
We also analyzed the DNA hydroxymethylation data by IPA and PANTHER programs in order to put the genes within biological contexts of potential relevance to neuronal plasticity, learning and memory and/or diseased states (Supplementary Figures S1 and S2). Biological processes with hyper-hydroxymethylated peaks in the SRvsCT comparison included biological adhesion, developmental process, localization and rhythmic process (Supplementary Figure S1). Molecular functions involved in addicted rats included transcription factor activity, receptor activity and transporter activity. Protein classes include receptors, cell adhesion molecules and calcium binding proteins. Several biological processes were also affected in the NAc of the nonaddicted rats (SSvsCT comparison) (Supplementary Figure S2). These include biological adhesion and immune system process. Peaks with increased DNA hydroxymethylation when comparing addicted with nonaddicted rats (SRvsSS) are located within or near genes that are transcription factors, receptors and transporters (Figure 3a). Peaks with decreased DNA hydroxymethylation in the addicted/nonaddicted comparison were also located in genes that encode for proteins that are transcription factors, receptors and transporters (Figure 3b).
Figure 4a shows the IPA results that confirm the role for some of these genes in potassium transport, cognition and synaptic plasticity. Indeed, several genes that code for potassium channels were found in the SSvsCT comparison. These include Kcnaj2 (intergenic L1), Kcnma1 (intron 4) and Kcnn2 (intergenic L1) that show increased DNA hydroxymethylation in the nonaddicted rats (Table 1). Similarly, the SRvsSS comparison also contained peaks in and around genes that code for potassium channels, including Kcna4 (intergenic), Kcnb2 (intergenic L1) and Kcnh1 (intron) that also show increased DNA hydroxymethylation, whereas Kcnd3 (intergenic L1) and Kcnt2 (intergenic L1) showed decreased hydroxymethylation (Table 1). Figures 4b and c show example of peaks located in intragenic regions of two genes coding for potassium channels.
Effects of methamphetamine self-administration (METH SA) on DNA hydroxymethylation and expression of potassium channels in the nucleus accumbens (NAc). (a) Ingenuity Pathway Analysis (IPA) shows that genes with differentially methylated peaks participate in transport of molecular, potassium transport and cognition. (b, c) UCSC (University of California, Santa Cruz) Genome Browser images of hydroxymethylated peaks in Kcnb2 and Kcnma1 intragenic sites, respectively. (d–k) mRNA expression of potassium channels in nonaddicted (shock-sensitive (SS)) and compulsive (shock-resistant (SR)) METH takers. **P<0.01, ***P<0.001 in comparison with control; #P<0.05, ##P<0.01, in comparison with SR.
Quantitative PCR of potassium channels
In order to investigate the impact of differential DNA hydroxymethylation on gene expression, we used quantitative PCR to measure the expression of several genes that code for potassium channels in the NAc. Figures 4d–k illustrate the results of METH SA on gene expression in the footshock-induced divergent phenotypes. Nonaddicted rats show increased expression of Kcna1 (Kv1.1) (F(2, 29)=5.05, P<0.013) and Kcna2 (Kv1.2) (F(2, 25)=8.81, P<0.001) expression in comparison with METH-addicted and control groups (Figures 4d and e, respectively). In addition, there were significant increases in the expression of Kcnab1 (also called Kcna1b, Kvb1) (F(2, 27)=5.51, P<0.01) in the nonaddicted group in comparison with the addicted rats (Figure 4f). Kcnb2 (Kv2.2) (F(2, 27)=7.5, P<0.0025) expression was also increased in the nonaddicted rats in comparison with control and compulsive METH takers (Figure 4g).
The nonaddicted rats also showed increased expression of other subtypes of potassium channels (Figures 4h–k). Kcnma1 (BK, SLO), coding for the large-conductance calcium-activated K+ channel subfamily M, α1, showed significant increased expression (F(2, 26)=5.21, P<0.013) in the nonaddicted rats in comparison with the control and the addicted rats (Figure 4h). Both Kcnn1 (SK1, KCa2.1) (F(2, 27)=5.70, P<0.0086), encoding for the small conductance Ca++ activated subfamily N α, member 1, and Kcnn2 (SK2, KCa2.2) (F(2, 26)=7.44, P<0.0028) showed increased mRNA levels in the nonaddicted rats in comparison with the control and METH-addicted rats (Figures 4i and j, respectively). Kcnn3 mRNA expression was not affected (Figure 4k).
In order to test for the specificity of the changes in gene expression, we also measured mRNA levels of potassium channels in the PFC and the dorsal striatum (Supplementary Figures S3 and S4). In the PFC, only Kcnn2 showed significant changes (F(2, 25)=3.43, P=0.0481) in mRNA levels, with the nonaddicted group showing ∼27% increases in comparison with the control group (Supplementary Figure S3). In the dorsal striatum, Kcna1 (F(2, 28)=8.82, P<0.0011) and Kcna2 (F(2, 29)=8.60, P<0.0012) showed significant decreases in both nonaddicted and addicted rats in comparison with the control group (Supplementary Figure S4), with there being no differences between the two METH-exposed rats. There were no significant changes in mRNA expression of any of the other potassium channels (Supplementary Figure S4). In general, these results indicate that the expression of potassium channels in the PFC and dorsal striatum did not help to differentiate compulsive METH takers from nonaddicted rats.
Discussion
METH addiction is a complex neuropsychiatric syndrome that is accompanied by pervasive changes in behaviors40 and a high rate of recidivism.41 The recent development of animal models that better mimic drug addiction in rodents29, 30, 42 should help to improve the characterization of cellular and molecular substrates of this complex behavioral syndrome and to usher the development of better pharmacological therapies. Here we show that compulsive METH taking is associated with large-scale changes in DNA hydroxymethylation in the rat NAc, consistent with a potential role for DNA hydroxymethylation processes in addiction.43 Genes that were affected in the METH-addicted rats included those that participate in calcium signaling, among others. In contrast, nonaddicted rats exhibited increased expression of mRNAs that code for several K+ channels in NAc, but not in the PFC and dorsal striatum.
K+ channels are a very large family of transmembrane proteins44, 45, 46 that regulate biological processes by controlling K+ flow across membrane pores47 and the firing properties of neurons.48, 49 Indeed, they play important roles in controlling the length of action potentials and stabilizing membrane potentials.48, 49 They are also involved in the regulation of diverse cellular processes including neurotransmitter release.50, 51 There are more than 100 mammalian genes that code for several subfamilies of K+ channels that are distributed throughout the brain and influence regional functions based on their codistribution with various neurotransmitter systems. Functional K+ channels consist of homo- and hetero-tetrameric structures composed of four α-subunits with a central core and auxiliary β-subunits.44, 46 K+ channels are located in somas, dendrites, spines and axon terminals where they can affect neuronal signaling based on their structural characteristics.
Voltage-gated potassium channels are differentially expressed in addicted and nonaddicted rats
In the present study, we have discovered that nonaddicted rats showed differential DNA hydroxymethylation and increased expression of two shaker genes, Kcna1 and Kcna2, that code for voltage-gated K+ (Kv) channels Kv1.1 and Kv1.2,44 respectively, in comparison with compulsive METH takers. Importantly, there were also increases in the expression of the accessory protein, KCNAB1, that codes for the potassium channel subunit-β152, 53 that forms heteromultimeric complexes with α-subunits such as Kv1.1 and Kv1.2.54 Association of α- and β-subunits in the brain increases the flexibility and diversity of potassium channels.54 Voltage-gated potassium channels are located on axons and axon terminals55 and serve to dampen neuronal excitability through the regulation of action potentials and the firing potentials of neurons.56 Interestingly, the shaker locus in drosophila that exhibit motoric behaviors was found to encode a potassium channel.57 Engineered Kcna1-null mice suffer from episodic eye blinking, increased startle response, hippocampal hyperexcitability and seizures.58 Deletion of Kcna2 also produces seizure activities in mice.59 Interestingly, Kcna1b-deficient mice showed normal synaptic plasticity in the hippocampus but impaired learning in the Morris water maze test.60 These data implicate a role of these K+ channels in the manifestation of complex behaviors. Thus, when taken together with the increased excitability reported in mice with Kv deletion, the increased expression of Kcna1 and Kcna2 mRNAs observed in the nonaddicted rats suggests the possibility that activation of these channels in the NAc might have served to suppress compulsive METH taking in the presence of adverse consequences. In addition to the increased expression of Kcna1 and Kcna2 in the nonaddicted rats, there were also significant increases in the mRNA levels of the shab-related delayed rectifier K+ channel, Kcnb2 (Kv2.2)61 in the nonaddicted rats in comparison with controls rats. Kv2.2 is highly expressed in the brain and is important in the regulation of somatodendritic excitability.49, 62 When taken together, the increased expression of voltage-gated potassium channels suggests that these channels might be coregulated to promote abstinence. Our data also hint to the potential use of activators of Kv channels63 in therapeutic interventions against METH addiction.
Increased expression of calcium-gated potassium channels in nonaddicted rats
We also discovered that the mRNA expression of 3 Ca++-activated K+ channels (Kcnma1, Kcnn1, and Kcnn2) was upregulated in the nonaddicted rats, suggesting that diverse potassium channels might work in concert to control or attenuate neuroadaptive mechanisms involved in the regulation of drug taking. Indeed, these calcium-activated channels are also found in subcellular locations such as soma, dendrites and axon terminals where they are known to regulate neuronal firing properties, neurotransmitter release, synaptic plasticity as well as learning and memory.46, 50, 64 It is thus possible to suggest that the increased expression in the nonaddicted animals may help to enhance mnemonic processes related to remembering adverse consequences associated with METH taking. The suggestion that these channels might be important to METH addiction is consistent with recent reports that have documented a role for these potassium channels in alcohol addiction.65, 66, 67 For example, chronic alcohol produced downregulation of surface SK2 (KCNN2) expression and reduced apamin-sensitive SK currents, whereas activation of SK2 channels by 1-ethyl-2-benzimidazolinone (1-EBIO) reduced alcohol intake.66 Interestingly, it has been reported that genetic data sets related to addiction contain families of potassium channels including KCNN1 for METH.67 Importantly, the authors also found that alcohol dependence was associated with decreased KCNN3 expression and blocking of KCNN3 activity increased alcohol intake. These reports are consistent with the fact that the SK channel activator, chlorzoxazone, can reduce excessive alcohol intake.68 Taken together with observations concerning alcohol addiction, our findings suggest that activators of SK channels69 might be important therapeutic targets for the treatment of substance use disorders.
Conclusion
In conclusion, quantitative analyses of DNA hydroxymethylation in METH-addicted and nonaddicted rats documented the presence of several differentially hydroxymethylated peaks in the NAc of these two phenotypes. Prominent among those are members of the classes of potassium channels including voltage- and calcium-activated channels that showed increased mRNA expression in the NAc of nonaddicted rats. These observations are consistent with data that had proposed a role of small-conductance calcium-gated potassium channels in alcohol addiction66 and voltage-gated channels in the effects of cocaine.70, 71 Thus, our experiments identify, for we believe the first time, a potential role for different classes of voltage-activated K+ channels in regulating abstinence and compulsive drug taking in a rat model of METH addiction. It is also important to note that Kv1 and Kv2 channels are differentially distributed on somatodendritic and axonal sites, suggesting the possibility that coregulation of these channels may, in part, serve to promote plastic changes that are responsible for the two dichotomous phenotypes. In any case, our large-scale quantitative analyses provide a rich source of information that can be mined for further assessments of specific roles that individual K+ channels might play in various aspects of rodent models of addiction. Finally, the present discoveries should help to usher novel drug development programs in the search for powerful medications in the therapeutic war against METH addiction.
References
Cadet JL, Brannock C, Jayanthi S, Krasnova IN . Transcriptional and epigenetic substrates of methamphetamine addiction and withdrawal: evidence from a long-access self-administration model in the rat. Mol Neurobiol 2015; 51: 696–717.
Krasnova IN, Chiflikyan M, Justinova Z, McCoy MT, Ladenheim B, Jayanthi S et al. CREB phosphorylation regulates striatal transcriptional responses in the self-administration model of methamphetamine addiction in the rat. Neurobiol Dis 2013; 58: 132–143.
Wright KN, Hollis F, Duclot F, Dossat AM, Strong CE, Francis TC et al. Methyl supplementation attenuates cocaine-seeking behaviors and cocaine-induced c-Fos activation in a DNA methylation-dependent manner. J Neurosci 2015; 35: 8948–8958.
Cadet JL, Brannock C, Ladenheim B, McCoy MT, Krasnova IN, Lehrmann E et al. Enhanced upregulation of CRH mRNA expression in the nucleus accumbens of male rats after a second injection of methamphetamine given thirty days later. PLoS One 2014; 9: e84665.
Everitt BJ . Neural and psychological mechanisms underlying compulsive drug seeking habits and drug memories–indications for novel treatments of addiction. Eur J Neurosci 2014; 40: 2163–2182.
Volkow ND, Wang GJ, Fowler JS, Tomasi D . Addiction circuitry in the human brain. Annu Rev Pharmacol Toxicol 2012; 52: 321–336.
Britt JP, Benaliouad F, McDevitt RA, Stuber GD, Wise RA, Bonci A . Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron 2012; 76: 790–803.
Parkes SL, Bradfield LA, Balleine BW . Interaction of insular cortex and ventral striatum mediates the effect of incentive memory on choice between goal-directed actions. J Neurosci 2015; 35: 6464–6471.
Cadet JL, Bisagno V . The primacy of cognition in the manifestations of substance use disorders. Front Neurol 2013; 4: 189.
Orsini CA, Moorman DE, Young JW, Setlow B, Floresco SB . Neural mechanisms regulating different forms of risk-related decision-making: insights from animal models. Neurosci Biobehav Rev 2015; 58: 147–167.
Bossert JM, Poles GC, Wihbey KA, Koya E, Shaham Y . Differential effects of blockade of dopamine D1-family receptors in nucleus accumbens core or shell on reinstatement of heroin seeking induced by contextual and discrete cues. J Neurosci 2007; 27: 12655–12663.
Fuchs RA, Evans KA, Parker MC, See RE . Differential involvement of the core and shell subregions of the nucleus accumbens in conditioned cue-induced reinstatement of cocaine seeking in rats. Psychopharmacology 2004; 176: 459–465.
Cornish JL, Duffy P, Kalivas PW . A role for nucleus accumbens glutamate transmission in the relapse to cocaine-seeking behavior. Neuroscience 1999; 93: 1359–1367.
Walsh JJ, Han MH . The heterogeneity of ventral tegmental area neurons: projection functions in a mood-related context. Neuroscience 2014; 282C: 101–108.
Berendse HW, Galis-de Graaf Y, Groenewegen HJ . Topographical organization and relationship with ventral striatal compartments of prefrontal corticostriatal projections in the rat. J Comp Neurol 1992; 316: 314–347.
Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol 2014; 15: R65.
Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y . Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci 2003; 23: 742–747.
Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M et al. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J Neurosci 2015; 35: 8042–8058.
Pierce RC, Wolf ME . Psychostimulant-induced neuroadaptations in nucleus accumbens AMPA receptor transmission. Cold Spring Harb Perspect Med 2013; 3: a012021.
Cadet JL, Bisagno V, Milroy CM . Neuropathology of substance use disorders. Acta Neuropathol 2014; 127: 91–107.
Jayanthi S, McCoy MT, Chen B, Britt JP, Kourrich S, Yau HJ et al. Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol Psychiatry 2014; 76: 47–56.
Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T et al. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One 2012; 7: e34236.
Whitfield TW Jr, Schlosburg JE, Wee S, Gould A, George O, Grant Y et al. kappa Opioid receptors in the nucleus accumbens shell mediate escalation of methamphetamine intake. J Neurosci 2015; 35: 4296–4305.
Cadet JL . Epigenetics of stress, addiction, and resilience: therapeutic implications. Mol Neurobiol 2016; 53: 545–560.
Godino A, Jayanthi S, Cadet JL . Epigenetic landscape of amphetamine and methamphetamine addiction in rodents. Epigenetics 2015; 10: 574–580.
Krasnova IN, Marchant NJ, Ladenheim B, McCoy MT, Panlilio LV, Bossert JM et al. Incubation of methamphetamine and palatable food craving after punishment-induced abstinence. Neuropsychopharmacology 2014; 39: 2008–2016.
Burman S . The challenge of sobriety: natural recovery without treatment and self-help groups. J Subst Abuse 1997; 9: 41–61.
Chen BT, Yau HJ, Hatch C, Kusumoto-Yoshida I, Cho SL, Hopf FW et al. Rescuing cocaine-induced prefrontal cortex hypoactivity prevents compulsive cocaine seeking. Nature 2013; 496: 359–362.
Pelloux Y, Murray JE, Everitt BJ . Differential vulnerability to the punishment of cocaine related behaviours: effects of locus of punishment, cocaine taking history and alternative reinforcer availability. Psychopharmacology 2015; 232: 125–134.
Vanderschuren LJ, Everitt BJ . Drug seeking becomes compulsive after prolonged cocaine self-administration. Science 2004; 305: 1017–1019.
Rubio FJ, Liu QR, Li X, Cruz FC, Leao RM, Warren BL et al. Context-induced reinstatement of methamphetamine seeking is associated with unique molecular alterations in Fos-expressing dorsolateral striatum neurons. J Neurosci 2015; 35: 5625–5639.
Krasnova IN, Justinova Z, Ladenheim B, Jayanthi S, McCoy MT, Barnes C et al. Methamphetamine self-administration is associated with persistent biochemical alterations in striatal and cortical dopaminergic terminals in the rat. PLoS One 2010; 5: e8790.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137.
Yang Y, Fear J, Hu J, Haecker I, Zhou L, Renne R et al. Leveraging biological replicates to improve analysis in ChIP-seq experiments. Comput Struct Biotechnol J 2014; 9: e201401002.
Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012; 22: 1813–1831.
Zhang Y, Lin YH, Johnson TD, Rozek LS, Sartor MA . PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics 2014; 30: 2568–2575.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589.
Mi H, Muruganujan A, Casagrande JT, Thomas PD . Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8: 1551–1566.
Richardson SR, Morell S, Faulkner GJ . L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet 2014; 48: 1–27.
Schulte MH, Cousijn J, den Uyl TE, Goudriaan AE, van den Brink W, Veltman DJ et al. Recovery of neurocognitive functions following sustained abstinence after substance dependence and implications for treatment. Clin Psychol Rev 2014; 34: 531–550.
Radfar SR, Rawson RA . Current research on methamphetamine: epidemiology, medical and psychiatric effects, treatment, and harm reduction efforts. Addict Health 2014; 6: 146–154.
Chen Y, Kramár EA, Chen LY, Babayan AH, Andres AL, Gall CM et al. Impairment of synaptic plasticity by the stress mediator CRH involves selective destruction of thin dendritic spines via RhoA signaling. Mol Psychiatry 2013; 18: 485–496.
Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci 2015; 18: 536–544.
Gutman GA, Chandy KG, Grissmer S, Lazdunski M, McKinnon D, Pardo LA et al. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev 2005; 57: 473–508.
Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y et al. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev 2005; 57: 509–526.
Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H . International Union of Pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol Rev 2005; 57: 463–472.
Tsantoulas C, McMahon SB . Opening paths to novel analgesics: the role of potassium channels in chronic pain. Trends Neurosci 2014; 37: 146–158.
Rasband MN . Clustered K+ channel complexes in axons. Neurosci Lett 2010; 486: 101–106.
Trimmer JS . Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron 2015; 85: 238–256.
Adelman JP, Maylie J, Sah P . Small-conductance Ca2+-activated K+ channels: form and function. Annu Rev Physiol 2012; 74: 245–269.
Kaczmarek LK . Non-conducting functions of voltage-gated ion channels. Nat Rev Neurosci 2006; 7: 761–771.
Leicher T, Roeper J, Weber K, Wang X, Pongs O . Structural and functional characterization of human potassium channel subunit beta 1 (KCNA1B). Neuropharmacology 1996; 35: 787–795.
Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO et al. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature 1994; 369: 289–294.
Pongs O, Leicher T, Berger M, Roeper J, Bahring R, Wray D et al. Functional and molecular aspects of voltage-gated K+ channel beta subunits. Ann NY Acad Sci 1999; 868: 344–355.
McKeown L, Swanton L, Robinson P, Jones OT . Surface expression and distribution of voltage-gated potassium channels in neurons (Review). Mol Membr Biol 2008; 25: 332–343.
Jan LY, Jan YN . Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol 2012; 590: 2591–2599.
Tempel BL, Papazian DM, Schwarz TL, Jan YN, Jan LY . Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. Science 1987; 237: 770–775.
Smart SL, Lopantsev V, Zhang CL, Robbins CA, Wang H, Chiu SY et al. Deletion of the K(V)1.1 potassium channel causes epilepsy in mice. Neuron 1998; 20: 809–819.
Brew HM, Gittelman JX, Silverstein RS, Hanks TD, Demas VP, Robinson LC et al. Seizures and reduced life span in mice lacking the potassium channel subunit Kv1.2, but hypoexcitability and enlarged Kv1 currents in auditory neurons. J Neurophysiol 2007; 98: 1501–1525.
Giese KP, Storm JF, Reuter D, Fedorov NB, Shao LR, Leicher T et al. Reduced K+ channel inactivation, spike broadening, and after-hyperpolarization in Kvbeta1.1-deficient mice with impaired learning. Learn Mem 1998; 5: 257–273.
Hwang PM, Glatt CE, Bredt DS, Yellen G, Snyder SH . A novel K+ channel with unique localizations in mammalian brain: molecular cloning and characterization. Neuron 1992; 8: 473–481.
Johnston J, Griffin SJ, Baker C, Skrzypiec A, Chernova T, Forsythe ID . Initial segment Kv2.2 channels mediate a slow delayed rectifier and maintain high frequency action potential firing in medial nucleus of the trapezoid body neurons. J Physiol 2008; 586: 3493–3509.
Wulff H, Castle NA, Pardo LA . Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov 2009; 8: 982–1001.
Sailer CA, Hu H, Kaufmann WA, Trieb M, Schwarzer C, Storm JF et al. Regional differences in distribution and functional expression of small-conductance Ca2+-activated K+ channels in rat brain. J Neurosci 2002; 22: 9698–9707.
Hopf FW, Bowers MS, Chang SJ, Chen BT, Martin M, Seif T et al. Reduced nucleus accumbens SK channel activity enhances alcohol seeking during abstinence. Neuron 2010; 65: 682–694.
Mulholland PJ, Becker HC, Woodward JJ, Chandler LJ . Small conductance calcium-activated potassium type 2 channels regulate alcohol-associated plasticity of glutamatergic synapses. Biol Psychiatry 2011; 69: 625–632.
Padula AE, Griffin WC 3rd, Lopez MF, Nimitvilai S, Cannady R, McGuier NS et al. KCNN genes that encode small-conductance Ca2+-activated K+ channels influence alcohol and drug addiction. Neuropsychopharmacology 2015; 40: 1928–1939.
Hopf FW, Simms JA, Chang SJ, Seif T, Bartlett SE, Bonci A . Chlorzoxazone, an SK-type potassium channel activator used in humans, reduces excessive alcohol intake in rats. Biol Psychiatry 2011; 69: 618–624.
Wulff H, Kolski-Andreaco A, Sankaranarayanan A, Sabatier JM, Shakkottai V . Modulators of small- and intermediate-conductance calcium-activated potassium channels and their therapeutic indications. Curr Med Chem 2007; 14: 1437–1457.
Kourrich S, Calu DJ, Bonci A . Intrinsic plasticity: an emerging player in addiction. Nat Rev Neurosci 2015; 16: 173–184.
Kourrich S, Hayashi T, Chuang JY, Tsai SY, Su TP, Bonci A . Dynamic interaction between sigma-1 receptor and Kv1.2 shapes neuronal and behavioral responses to cocaine. Cell 2013; 152: 236–247.
Acknowledgements
This work was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA.
Author contributions
JLC, the corresponding author, supervised all aspects of the experiment, performed IPA analysis and wrote the manuscript. CB performed IPA and PANTHER analyses of the data. INK wrote the animal protocol and performed the self-administration experiment. SJ performed the hMeDIP assay and contributed to the writing of the paper. BL, MTM and DW performed quantitative PCR analyses. BL and MTM also dissected rat brains and provided technical assistance with the self-administration experiment. AG assisted with data analysis. MP performed the bioinformatics analyses on the hMeDIP sequencing data. RSL performed the hMeDIP sequencing experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Cadet, J., Brannock, C., Krasnova, I. et al. Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol Psychiatry 22, 1196–1204 (2017). https://doi.org/10.1038/mp.2016.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.48
This article is cited by
-
Incubation of methamphetamine craving in punishment-resistant individuals is associated with activation of specific gene networks in the rat dorsal striatum
Molecular Psychiatry (2024)
-
Deep sequencing of candidate genes identified 14 variants associated with smoking abstinence in an ethnically diverse sample
Scientific Reports (2024)
-
Footshock-Induced Abstinence from Compulsive Methamphetamine Self-administration in Rat Model Is Accompanied by Increased Hippocampal Expression of Cannabinoid Receptors (CB1 and CB2)
Molecular Neurobiology (2022)
-
Antidepressant activity of pharmacological and genetic deactivation of the small-conductance calcium-activated potassium channel subtype-3
Psychopharmacology (2022)
-
Behavioral sensitization induced by methamphetamine causes differential alterations in gene expression and histone acetylation of the prefrontal cortex in rats
BMC Neuroscience (2021)