Abstract
The prognostic role of MYC has been well documented in non-central nervous system diffuse large B-cell lymphoma; however, it remains controversial in central nervous system diffuse large B-cell lymphoma. To investigate the prognostic value of MYC, we analyzed the MYC protein expression by immunohistochemistry, mRNA expression by RNA in situ hybridization, and gene status by fluorescence in situ hybridization in 74 cases of central nervous system diffuse large B-cell lymphoma. Moreover, we examined the correlation between MYC translocation, mRNA expression, and protein expression. The mean percentage of MYC immunopositive cells was 49%. Using a 44% cutoff value, 49 (66%) cases showed MYC protein overexpression. The result of mRNA in situ hybridization using the RNA scope technology was obtained using the H-scoring system; the median value was 34.2. Using the cutoff value of 63.5, 16 (22%) cases showed MYC mRNA overexpression. MYC gene rearrangement was detected in five out of 68 (7%) cases. MYC translocation showed no statistically significant correlation with mRNA expression; however, all MYC translocation-positive cases showed MYC protein overexpression, with a higher mean percentage of MYC protein expression than that of translocation-negative cases (78 vs 48%, P=0.001). The level of MYC mRNA expression was moderately correlated with the level of MYC protein expression (P<0.001). The mean percentage of MYC protein expression in the high MYC mRNA group was higher than that in the low MYC mRNA group (70 vs 47%, P<0.001). A univariate analysis showed that age over 60 years, Eastern Cooperative Oncology Group (ECOG) performance status ≥2 and MYC protein overexpression were significantly associated with an increased risk of death. MYC translocation and MYC mRNA expression had no prognostic significance. On multivariate analysis, MYC protein overexpression and ECOG score retained prognostic significance.
Similar content being viewed by others
Main
Primary central nervous system diffuse large B-cell lymphoma is a rare form of diffuse large B-cell lymphoma with an intracerebral or intraocular location that accounts for 1% of all non-Hodgkin lymphomas.1 The tumor cells of central nervous system diffuse large B-cell lymphoma are relatively homogeneous and are characterized by a centroblastic appearance, a late germinal center B-cell origin (as assessed using gene expression profiling), and a non-germinal center B-cell phenotype (as assessed using immunohistochemistry based on the Hans algorithm).2, 3, 4 The prognosis of central nervous system diffuse large B-cell lymphoma is generally unfavorable, because of the unique tumor cell biology, location itself, or central nervous system-specific microenvironment. However, prognostic markers for this condition have not been identified.
In recent years, the role of the MYC gene in the prognosis of non-central nervous system diffuse large B-cell lymphoma has been elucidated. MYC is a transcription factor that activates many genes involved in diverse cellular processes, including the regulation of cell size, survival, proliferation, metabolism, and angiogenesis. Deregulated MYC expression is involved in the pathogenesis of various solid tumor and lymphoid malignancies.5 MYC rearrangements have been identified in 5–15% of non-central nervous system diffuse large B-cell lymphomas and are associated with a poor prognosis, especially when combined with BCL2 and/or BCL6 rearrangement ('double-hit' and 'triple-hit' lymphomas).6, 7, 8, 9 In addition, high MYC and/or BCL2 protein expression results in an adverse prognosis, regardless of the presence of MYC rearrangements.10, 11, 12, 13, 14
MYC rearrangement in central nervous system diffuse large B-cell lymphoma has been reported recently, at a lower incidence (0–9%) than that in non-central nervous system diffuse large B-cell lymphoma. There are limited data on the prognostic significance of MYC rearrangement in central nervous system diffuse large B-cell lymphoma.15, 16, 17, 18 In contrast, high MYC protein expression (43–92%) in central nervous system diffuse large B-cell lymphoma has been reported in recent studies with an inconsistent association with overall survival.17, 18, 19 The mechanisms underlying the deregulation of MYC expression in central nervous system diffuse large B-cell lymphoma have been uncovered. Although aberrant somatic hypermutation involving the MYC gene has been detected in central nervous system diffuse large B-cell lymphoma, its clinical significance is unknown at present.20 Recent gene expression profiling analyses to identify genes that are differentially expressed between central nervous system diffuse large B-cell lymphoma and non-central nervous system diffuse large B-cell lymphoma did not identify the MYC gene.2, 21
In the current study, we investigated the association between MYC protein expression, MYC gene rearrangement, and MYC mRNA expression, as well as their prognostic value for central nervous system diffuse large B-cell lymphoma.
Materials and methods
Sample Collection and Processing
Seventy-four pretreatment tumor biopsies of patients diagnosed with de novo central nervous system diffuse large B-cell lymphoma according to the WHO classification (2008) criteria were collected.1 Patients were diagnosed between January 1995 and December 2012 at the Department of Pathology of the Samsung Medical Center (Seoul, Korea). Clinical information was collected from the medical records and included age, performance status according to the Eastern Cooperative Oncology Group (ECOG), serum lactate dehydrogenase levels at the time of diagnosis, sex, type of treatment, and date of the last follow-up or death. The study was approved by the research ethics boards of our institutions, according to the Declaration of Helsinki.
Tissue Microarrays and Immunohistochemistry
Formalin-fixed, paraffin-embedded specimens were used to generate a tissue microarray. Representative 1 mm cores of each case, in duplicate, were taken from tissue blocks. Immunohistochemical stains of 4-μm paraffin sections of the tissue microarray blocks were performed using a Bond Max automated immunostainer (Leica Biosystems, Melbourne, Australia). Monoclonal antibodies against CD20 (L26, 1/200; Dako, Glostrup, Denmark), CD3 (polyclonal, 1/200; Dako), CD10 (56C6, 1/250; Novocastra, Newcastle upon Tyne, UK), BCL6 (LN22, 1/80; Novocastra), MUM1 (MUM1p, 1/500; Dako), BCL2 (124, 1/100; Dako), and MYC (Y69, cat:ab32072, 1/100; Abcam, Burlingame, CA, USA) were used. The cell-of-origin subtype was determined using the Hans algorithm.4 MYC immunoreactivity was evaluated manually on an average of 200 cells per case. MYC and BCL2 positivity was analyzed using the X-tile statistical software (http://www.tissuearray.org/rimmlab) to determine the optimum cutoff point for dichotomizing the expression of the MYC protein (≥44%) and BCL2 protein (≥70%) based on patient survival.22 The reproducibility of MYC and BCL2 protein expression measurements by IHC was checked by comparing the results obtained by two pathologists (SMS and YHK).
MYC mRNA In situ Hybridization Assay
The MYC mRNA expression was examined in the 74 patients by RNA in situ hybridization using the RNA scope technology (Advanced Cell Diagnostics, Hayward, CA, USA). All RNA in situ hybridization slides were captured and were printed as hard copies. The RNA signals were evaluated by two independent observers (SMS and YHK). MYC mRNA molecules were detected with single-copy detection sensitivity. Single-molecule signals were quantified on a cell-by-cell basis by manual counting. The signals per cell were divided into five groups (0 dots/cell, 1–3 dots/cell, 4–9 dots/cell, 10–15 dots/cell, and>15 dots/cell with >10% of dots in clusters). Each sample was evaluated for the percentage of cells in each group. The H-score was calculated by adding up the percentage of cells in each group, with a weight assigned to each group, using the formula provided by the manufacturer (Supplementary Table 1). H-scores are given on a scale of 0–400.
Fluorescence In situ Hybridization (FISH) Assay
MYC gene rearrangements were evaluated by FISH using a Vysis LSI MYC dual color, break-apart probe (Abbott Molecular, Abbott Park, IL, USA).23 BCL2 gene rearrangements were investigated using a Vysis LSI BCL2 dual color, break-apart probe (Abbott Molecular). Cases with break-apart signals in >3% of nuclei were considered positive for the presence of a translocation.
Statistical Anlaysis
The baseline characteristics are described as the mean and s.d., or median and range, for quantitative variables; and as frequency and percentage for categorical variables. The analysis of the correlation between clinicopathological features and the results of MYC RNA in situ hybridization and MYC immunohistochemistry was performed using the χ2 and Fisher’s exact test. Student’s t-test and the Mann–Whitney U-test were used when applicable for comparisons among study groups. Pearson’s correlation analysis (r) was used to assess the linear relationship between the MYC immunopositive tumor cell percentage and MYC mRNA expression score based on the H-scoring system. Survival curves were plotted according to the Kaplan–Meier method and were compared using the log-rank test. The Cox proportional hazard model was used for univariate and multivariate survival analyses. A multivariate analysis was performed including the variables that were found to be predictive by univariate analysis (P<0.05). Overall survival was defined as the time between the date of the diagnostic biopsy and the date of death or of the last follow-up. Statistical analyses were carried out using the SPSS statistical package, version 23.0 (IBM, NY, USA). Statistical significance was set at P<0.05 (two sided).
Results
Patient Characteristics
The study cohort consisted of 74 immunocompetent patients with central nervous system diffuse large B-cell lymphoma, whose characteristics are summarized in Table 1. The overall sex composition was 40 (54%) males and 34 (46%) females. The median age of the patients at diagnosis was 58 years (range, 19–82 years). Fifty-seven out of 74 (77%) patients had an ECOG performance status score ≥2. Lactate dehydrogenase levels at diagnosis were elevated in 20 out of the 68 (29%) patients with available data. An International Prognostic Index (IPI) of more than high-intermediate risk was observed in 11 out of 68 (16%) patients. Seventy-two of the 74 (97%) patients had available treatment data. Sixty-nine out of 72 (96%) patients received methotrexate-based chemotherapy: high-dose methotrexate alone (n=5), high-dose methotrexate with cytarabine (n=3), high-dose methotrexate with rituximab and cytarabine (n=1), high-dose methotrexate with vincristine (n=6), high-dose methotrexate with vincristine and cytarabine (n=8), high-dose methotrexate with procarbazine and vincristine (n=4), high-dose methotrexate with procarbazine and vincristine with cytarabine (n=41), and high-dose methotrexate with procarbazine and vincristine with rituximab (n=1). Whole-brain radiation therapy (WBRT) was added to chemotherapy in 56 patients. One patient was treated with WBRT only and two patients received palliative care only.
Of the treated patients with follow-up information, the median follow-up was 35.2 months (range, 1.8–148.8 months) and the median overall survival time was 61.8 months (95% confidence interval, 40.8–94.2). Lymphoma was the cause of death in all the patients who were followed until death. Nine patients (12%) were lost to the follow-up and 45 patients (61%) had died at the time of data collection.
Morphology and Immunohistochemistry
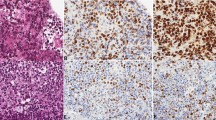
All 74 cases were positive for CD20 and negative for CD3. Fifty-nine out of 74 cases (80%) were negative for CD10 and positive for both BCL6 and MUM1; these cases were of the non-germinal center type according to the Hans algorithm (Figure 1a). The other 15 cases were of the germinal center type by the Hans algorithm; nine were negative for both CD10 and MUM1 and positive for BCL6, four were positive for all three markers, and two were negative for MUM1 and positive for both CD10 and BCL6. BCL2 was expressed in 54 out of 74 cases (73%).
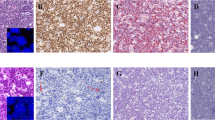
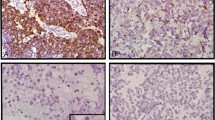
Representative example of primary central nervous system diffuse large B-cell lymphoma. (a) Hematoxylin and eosin staining showing large neoplastic cells with prominent nucleoli (original magnification, × 400). (b) MYC staining showing a high percentage of cells with positive protein expression (original magnification, × 400). (c) RNA in situ hybridization analysis showing positive MYC mRNA expression (original magnification, 400 × ). (d) MYC rearrangement shown by FISH (one split signal, red and green, and one fused signal). FISH, fluorescence in situ hybridization.
The mean percentage of MYC immunopositive cells was 49% (95% confidence interval, 44–54%) and the median value was 52% (range, 1–90%). MYC protein expression was exclusively nuclear in all cases (Figure 1b). Using the cutoff value of 44%, as assessed using the X-tile package, 49 out of 74 cases (66%) were considered MYC immunopositive cases. The relationships between MYC protein expression and clinicopathological findings are shown in Table 1. Patients with MYC protein overexpression were more frequently female and more frequently showed high-risk IPI score, non-germinal center phenotype, MUM1 protein expression, and MYC mRNA expression. The proportion of MYC immunopositive cases was higher for the non-germinal center than for the germinal center phenotype (75 vs 33%, respectively, P=0.003) (Pearson’s χ2-test). The mean percentage of MYC immunopositive cells was higher in the non-germinal center than in the germinal center phenotype (52 vs 36%, respectively, P=0.01) (Student’s t-test).
Determination of MYC mRNA Expression via RNA in situ Hybridization
To assess MYC mRNA expression levels, a quantitative RNA in situ hybridization analysis was performed by calculating the H-score according to the ;manufacturer’s guidelines (Supplementary Table 1). The mean MYC mRNA H-score was 45.3 (95% confidence interval, 34.8–55.8) and the median value was 34.2 (range, 1.0–207.5). Using the cutoff value of 63.5, as determined using the X-tile package, 16 out of 74 cases (22%) were classified as high MYC mRNA cases (Figure 1c). The relationships between MYC mRNA level and clinicopathological findings are shown in Table 1. The proportion of high MYC mRNA cases was greater in MYC immunopositive cases than it was in MYC immunonegative cases (29 vs 8%, respectively, P=0.042) (Pearson’s χ2-test). The level of MYC mRNA expression was moderately correlated with the level of MYC protein expression (P<0.001, r=0.544) (Pearson’s correlation analysis) (Figure 2a). The level of MYC protein expression in the high MYC mRNA group was higher than that in the low-MYC mRNA group (Figure 2b) (median, 70 vs 47%, respectively; P<0.001) (Mann–Whitney U-test).
FISH Analyses
FISH assays to determine MYC aberrations were performed in all cases, and informative FISH analyses were obtained in 68 out of 74 (92%) cases. Five (7%) cases showed MYC translocation (Figure 1d). No other genetic aberrations, including amplification or polysomy, were observed. In addition, BCL2 FISH was performed in the five MYC-FISH-positive cases, and no translocation involving BCL2 was found. All cases of MYC translocation belonged to the MYC immunopositive group. The mean percentage of MYC immunopositive cells in the MYC-FISH-positive group was higher than that in the MYC-FISH-negative group (78 vs 47%, respectively, P=0.001) (Student’s t-test) (Figure 3a). However, MYC mRNA expression levels were not significantly different between the MYC-FISH-positive and MYC-FISH-negative cases (median, 110.6 vs 34.6, respectively; P=0.084) (Mann–Whitney U-test) (Figure 3b). The five MYC-FISH-positive cases were divided into two low MYC RNA cases and three high MYC RNA cases (Table 1).
Factors Associated with Clinical Outcome
Table 2 presents the results of univariate and multivariate survival analyses that included age, sex, ECOG status, cell-of-origin phenotype according to the Hans algorithm, BCL6 protein expression, BCL2 protein expression, MYC protein expression, MYC and BCL2 protein co-expression, and MYC mRNA expression status. A univariate analysis showed that age over 60 years, ECOG≥2, and MYC immunopositivity were significantly associated with an increased risk of death. However, there were no significant differences in survival according to MYC mRNA expression status (Table 2 and Figure 4) (log-rank test). The 3-year overall survival rate was not different between the high-MYC mRNA group and the low-MYC mRNA group (53 vs 66%, P=0.352). On multivariate analysis, MYC protein expression and ECOG retained prognostic significance (P=0.016 and P=0.004, respectively). The prognostic significance of MYC protein expression and MYC mRNA expression was evaluated according the cutoff points determined using the X-tile package; cutoffs at 44% and 63.5 points, respectively, exhibited the best correlation with survival. As informative MYC FISH analysis was possible in 68 out of 74 cases, the prognostic evaluation was applied to these 68 cases only; there were no significant differences in survival according to MYC translocation status (P=0.503) (log-rank test).
Overall survival of patients with primary central nervous system diffuse large B-cell lymphoma. Kaplan–Meier curves represent overall survival according to (a) high MYC protein expression and (b) high MYC mRNA expression. Log-rank tests for a and b yielded P=0.016 and P=0.055, respectively. Total evaluable patients for the analyses: (a) n=74 and (b) n=74.
Discussion
Primary lymphoma of the central nervous system is a specific type of diffuse large B-cell lymphoma that is confined to the central nervous system. The central nervous system is an immunoprivileged organ without a classical lymphatic drainage system, which creates a specific microenvironment for developing lymphoid malignancy. The three major components of a central nervous system with primary central nervous system lymphoma are malignant B cells, resident brain cells, and reactive inflammatory bystander cells.3 The interaction between the tumor B cells, astrocytes, microglial cells, reactive T cells, and endothelial cells via the expression of chemokines, cytokines, cell-adhesion molecules, osteopontin, and major histocompatibility complex class II antigen may contribute to the characteristic perivascular tumor cell cuffing and diffuse brain infiltration. However, at present, the mechanisms underlying the selective tropism and confinement to the central nervous system are unclear.3, 24, 25, 26, 27
A variety of prognostic markers have been investigated in central nervous system diffuse large B-cell lymphoma. A perivascular tumor cell infiltration pattern was identified as a poor prognostic factor.28 The presence of reactive perivascular T cells was associated with superior overall survival.26, 28 Microvessel density appeared to be an inconsistent prognostic factor in central nervous system diffuse large B-cell lymphoma.29, 30, 31 BCL6 expression has been reported to have an inconsistent association with prognosis.17, 18, 32, 33, 34, 35 Chromosome 6q deletions and homozygous 9p deletions were associated with inferior progression-free survival and overall survival.15, 36
In this work, we evaluated MYC as a prognostic marker for central nervous system diffuse large B-cell lymphoma. MYC translocation has a lower prevalence (0–9%) in central nervous system diffuse large B-cell lymphoma compared with non-central nervous system diffuse large B-cell lymphoma.15, 16, 17, 18 MYC protein overexpression is more frequent (43–92%) than MYC translocation in central nervous system diffuse large B-cell lymphoma.16, 17, 18, 34 The prognostic value of MYC protein expression for central nervous system diffuse large B-cell lymphoma remains controversial. Therefore, we analyzed MYC protein expression by immunohistochemistry, MYC gene status by FISH, and MYC mRNA expression by RNA in situ hybridization, to investigate the correlation between these parameters and to evaluate their prognostic significance.
MYC protein overexpression was more frequent in central nervous system diffuse large B-cell lymphoma (43–92%) than it was in non-central nervous system diffuse large B-cell lymphoma (29–64%).10, 11, 12, 14, 16, 17, 18, 34 MYC protein expression was identified in 66% of our cohort. The high prevalence of MYC protein overexpression observed in central nervous system diffuse large B-cell lymphoma may be related to the predominance of the non-germinal center subtype in central nervous system diffuse large B-cell lymphoma, as MYC expression is more frequent in the non-germinal center subtype of non-central nervous system diffuse large B-cell lymphoma.7, 10 Our data also showed the presence of a significant association between MYC protein overexpression and the non-germinal center subtype (P=0.003, Table 1).
The data regarding the prognostic value of MYC protein expression for central nervous system diffuse large B-cell lymphoma are inconsistent.17, 18, 19 Two studies comprising 14 and 42 central nervous system diffuse large B-cell lymphoma cases performed by Chang et al.19 and Tapia et al.,18 respectively, reported that MYC overexpression was associated with an adverse prognosis; however, in the cohort of 59 patients reported by Gill et al.,17 no association was found with clinical outcome. In our work, MYC protein expression was related to poor overall survival (P=0.016) and appeared as an independent prognostic factor in a multivariate analysis (P=0.016). The impact of MYC and BCL2 co-expression was also evaluated as their unfavorable clinical impact was reported in non-central nervous system diffuse large B-cell lymphoma;7, 11, 13 however, concurrent expression of MYC and BCL2 was not of prognostic value in this work, similar to the study of Tapia et al.18 In addition, a prognostic difference according to germinal center/non-germinal center subtype was not observed in this study, which was in line with the studies of Hattab et al.39 and Raoux et al.40
We also evaluated MYC mRNA expression using RNA scope. The central nervous system diffuse large B-cell lymphoma showed high-MYC mRNA expression, which exceeds that reported for non-central nervous system diffuse large B-cell lymphoma.14 The impact of MYC mRNA expression on patient outcome was not statistically significant (P=0.055).
MYC translocation was observed only in 7% of cases. There was no significant difference in prognosis according to MYC translocation status. Consistent with a previous study, the rarity of MYC translocation found here makes it unsuitable as a prognostic tool for central nervous system diffuse large B-cell lymphoma.15, 16, 17, 18 Furthermore, BCL2 FISH was performed in the five MYC-FISH-positive cases to investigating additional gene alterations, and no translocation involving BCL2 was detected.
Deregulation of the MYC oncogene contributes to tumorigenesis, via its transcriptional and nontranscriptional roles in cellular processes, such as proliferation, differentiation, and metabolism. MYC deregulation is mediated not only by direct alterations, such as amplification or chromosomal translocation, but also by the activation of many receptor signaling pathways. The deregulated expression of MYC results in an increase of MYC protein expression.5 The genetic mechanism underlying MYC protein expression in central nervous system diffuse large B-cell lymphoma has not been elucidated. However, studies aimed at disclosing the genetic and epigenetic mechanisms of MYC regulation are in progress. Aberrant somatic hypermutation of the MYC gene has been identified in nine out of 10 cases of central nervous system diffuse large B-cell lymphoma; however, the impact of MYC mutations on MYC protein expression in central nervous system diffuse large B-cell lymphoma remains unknown.20
We examined the correlation between MYC translocation, MYC mRNA expression, and MYC protein expression. In our study, MYC translocation showed no statistically significant correlation with MYC mRNA expression (P=0.081) (Fisher’s exact test). However, all cases with MYC translocation showed MYC protein overexpression. These results suggest that MYC translocation leads to overexpression of the MYC protein, although many more MYC-translocation-negative cases overexpress MYC protein via other mechanisms. The mechanisms of MYC deregulation are not restricted to translocations or amplifications of the MYC locus, and MYC can be deregulated by any one of several mechanisms that target its expression and/or activity, either directly or indirectly.41
In our study, MYC mRNA expression showed moderate positive correlation with MYC protein expression (P<0.001, r=0.544) (Pearson’s correlation analysis). Several reports have shown more frequent MYC protein overexpression in central nervous system diffuse large B-cell lymphoma than in non-central nervous system diffuse large B-cell lymphoma,10, 11, 12, 14 while the difference of MYC mRNA expression between them was controversial. Rubenstein et al.27 presented increased MYC mRNA expression in central nervous system diffuse large B-cell lymphoma compared to non-central nervous system diffuse large B-cell lymphoma. Brunn et al.16 reported that MYC mRNA level in central nervous system diffuse large B-cell lymphoma was not different from that of non-central nervous system diffuse large B-cell. Our results suggest that MYC protein overexpression cannot be completely accounted for by mRNA overexpression; posttranscriptional or posttranslational modulation may also play a role.
The MYC protein is highly unstable and its destruction is mediated by post-translational modification, ubiquitination, and degradation.42, 43 Mutation of the MYC gene may stabilize the MYC protein and contribute to tumorigenesis, as in Burkitt’s lymphoma.38, 44, 45 Moreover, deregulation of microRNAs (miRNAs) may be associated with MYC protein overexpression, via translational modulation. Numerous miRNAs have been shown to regulate MYC expression.46, 47 MYC-regulated miRNAs, such as miR-17-5p and miR-20a, which belong to the miR-17-92 clusters, are upregulated in central nervous system diffuse large B-cell lymphoma compared with non-central nervous system diffuse large B-cell lymphoma.48 MYC-activated miR-17-92 clusters contribute to lymphomagenesis via amplification of B-cell receptor (BCR) signaling, and stimulated BCR responses result in elevation of MYC itself, thus forming a feed-forward loop.49 Further studies of miRNA-mediated pathways in central nervous system diffuse large B-cell lymphoma may explain the observed MYC overexpression.
We evaluated MYC mRNA expression using the RNA scope method, which quantifies RNA via an in situ detection method. This technique, which was first developed by Wang et al.,37 enables direct counting of mRNA molecules in single cells in routine formalin-fixed, paraffin-embedded tissue specimens using bright-field microscopy. The false-negative results obtained from admixtures of many non-malignant cells with tumor cells in real-time RT–PCR can be overcome using this method; however, specimen quality may be another problematic issue. In our study, the experiment was successfully performed using paraffin archives and manual counting of a single signal, followed by H-scoring, which yields accurate measurement of mRNA expression.
In summary, we describe MYC protein expression as an important prognostic marker for the stratification of patients with central nervous system diffuse large B-cell lymphoma. Moreover, we evaluated MYC mRNA expression using the RNA in situ hybridization method, and assessed its impact on patient survival for the first time. In addition, we assessed the correlation between MYC gene status, MYC mRNA expression, and MYC protein expression in central nervous system diffuse large B-cell lymphoma. The present study had several limitations. Because of its retrospective nature, the patients were not highly selected, as would be done in a prospective study. Further studies of the complete molecular mechanisms underlying MYC protein overexpression would be useful to validate and expand our findings.
References
Kluin PM, Deckert M, Ferry JA, Primary diffuse large B-cell lymphoma of the CNS. In: Swerdlow SH, Campo E, Harris NL, et al. (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. IARC Press: Lyon, France, 2008, pp 240–241..
Montesinos-Rongen M, Brunn A, Bentink S et al, Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia 2008;22:400–405.
Deckert M, Montesinos-Rongen M, Brunn A et al, Systems biology of primary CNS lymphoma: from genetic aberrations to modeling in mice. Acta Neuropathol 2014;127:175–188.
Hans CP, Weisenburger DD, Greiner TC et al, Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–282.
Dang CV . MYC on the path to cancer. Cell 2012;149:22–35.
Savage KJ, Johnson NA, Ben-Neriah S et al, MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009;114:3533–3537.
Johnson NA, Savage KJ, Ludkovski O et al, Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood 2009;114:2273–2279.
Snuderl M, Kolman OK, Chen YB et al, B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol 2010;34:327–340.
Akyurek N, Uner A, Benekli M et al, Prognostic significance of MYC, BCL2, and BCL6 rearrangements in patients with diffuse large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone plus rituximab. Cancer 2012;118:4173–4183.
Hu S, Xu-Monette ZY, Tzankov A et al, MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013;121:4021–4031, quiz 4250.
Horn H, Ziepert M, Becher C et al, MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013;121:2253–2263.
Valera A, Lopez-Guillermo A, Cardesa-Salzmann T et al, MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica 2013;98:1554–1562.
Green TM, Young KH, Visco C et al, Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012;30:3460–3467.
Johnson NA, Slack GW, Savage KJ et al, Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012;30:3452–3459.
Cady FM, O'Neill BP, Law ME et al, Del(6)(q22) and BCL6 rearrangements in primary CNS lymphoma are indicators of an aggressive clinical course. J Clin Oncol 2008;26:4814–4819.
Brunn A, Nagel I, Montesinos-Rongen M et al, Frequent triple-hit expression of MYC, BCL2, and BCL6 in primary lymphoma of the central nervous system and absence of a favorable MYC(low)BCL2 (low) subgroup may underlie the inferior prognosis as compared to systemic diffuse large B cell lymphomas. Acta Neuropathol 2013;126:603–605.
Gill KZ, Iwamoto F, Allen A et al, MYC protein expression in primary diffuse large B-cell lymphoma of the central nervous system. PLoS One 2014;9:e114398.
Tapia G, Baptista MJ, Munoz-Marmol AM et al, MYC protein expression is associated with poor prognosis in primary diffuse large B-cell lymphoma of the central nervous system. APMIS 2015;123:596–603.
Chang CC, Kampalath B, Schultz C et al, Expression of p53, c-Myc, or Bcl-6 suggests a poor prognosis in primary central nervous system diffuse large B-cell lymphoma among immunocompetent individuals. Arch Pathol Lab Med 2003;127:208–212.
Montesinos-Rongen M, Van Roost D, Schaller C et al, Primary diffuse large B-cell lymphomas of the central nervous system are targeted by aberrant somatic hypermutation. Blood 2004;103:1869–1875.
Lim, do H, Kim WS, Kim SJ et al, Microarray gene-expression profiling analysis comparing PCNSL and non-CNS diffuse large B-cell lymphoma. Anticancer Res 2015;35:3333–3340.
Camp RL, Dolled-Filhart M, Rimm DL . X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252–7259.
Munoz-Marmol AM, Sanz C, Tapia G et al, MYC status determination in aggressive B-cell lymphoma: the impact of FISH probe selection. Histopathology 2013;63:418–424.
Brunn A, Montesinos-Rongen M, Strack A et al, Expression pattern and cellular sources of chemokines in primary central nervous system lymphoma. Acta Neuropathol 2007;114:271–276.
Kadoch C, Dinca EB, Voicu R et al, Pathologic correlates of primary central nervous system lymphoma defined in an orthotopic xenograft model. Clin Cancer Res 2009;15:1989–1997.
Ponzoni M, Berger F, Chassagne-Clement C et al, Reactive perivascular T-cell infiltrate predicts survival in primary central nervous system B-cell lymphomas. Br J Haematol 2007;138:316–323.
Rubenstein JL, Fridlyand J, Shen A et al, Gene expression and angiotropism in primary CNS lymphoma. Blood 2006;107:3716–3723.
He M, Zuo C, Wang J et al, Prognostic significance of the aggregative perivascular growth pattern of tumor cells in primary central nervous system diffuse large B-cell lymphoma. Neuro Oncol 2013;15:727–734.
Takeuchi H, Matsuda K, Kitai R et al, Angiogenesis in primary central nervous system lymphoma (PCNSL). J Neurooncol 2007;84:141–145.
Sugita Y, Takase Y, Mori D et al, Endoglin (CD 105) is expressed on endothelial cells in the primary central nervous system lymphomas and correlates with survival. J Neurooncol 2007;82:249–256.
D'Haene N, Catteau X, Maris C et al, Endothelial hyperplasia and endothelial galectin-3 expression are prognostic factors in primary central nervous system lymphomas. Br J Haematol 2008;140:402–410.
Braaten KM, Betensky RA, de Leval L et al, BCL-6 expression predicts improved survival in patients with primary central nervous system lymphoma. Clin Cancer Res 2003;9:1063–1069.
Lossos C, Bayraktar S, Weinzierl E et al, LMO2 and BCL6 are associated with improved survival in primary central nervous system lymphoma. Br J Haematol 2014;165:640–648.
Rubenstein JL, Hsi ED, Johnson JL et al, Intensive chemotherapy and immunotherapy in patients with newly diagnosed primary CNS lymphoma: CALGB 50202 (Alliance 50202). J Clin Oncol 2013;31:3061–3068.
Camilleri-Broet S, Criniere E, Broet P et al, A uniform activated B-cell-like immunophenotype might explain the poor prognosis of primary central nervous system lymphomas: analysis of 83 cases. Blood 2006;107:190–196.
Gonzalez-Aguilar A, Idbaih A, Boisselier B et al, Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res 2012;18:5203–5211.
Wang F, Flanagan J, Su N et al, RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–29.
Thomas LR, Tansey WP . Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res 2011;110:77–106.
Hattab EM, Martin SE, Al-Khatib SM et al, Most primary central nervous system diffuse large B-cell lymphomas occurring in immunocompetent individuals belong to the nongerminal center subtype: a retrospective analysis of 31 cases. Mod Pathol 2010;23:235–243.
Raoux D, Duband S, Forest F et al, Primary central nervous system lymphoma: immunohistochemical profile and prognostic significance. Neuropathology 2010;30:232–240.
Meyer N, Penn LZ . Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976–990.
Gregory MA, Qi Y, Hann SR . Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 2003;278:51606–51612.
Gregory MA, Hann SR . c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 2000;20:2423–2435.
Salghetti SE, Kim SY, Tansey WP . Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J 1999;18:717–726.
Wang X, Cunningham M, Zhang X et al, Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res 2011;71:925–936.
Psathas JN, Thomas-Tikhonenko A . MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med 2014;4.
Sachdeva M, Zhu S, Wu F et al, p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 2009;106:3207–3212.
Fischer L, Hummel M, Korfel A et al, Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol 2011;13:1090–1098.
Psathas JN, Doonan PJ, Raman P et al, The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood 2013;122:4220–4229.
Acknowledgements
This study was supported by a grant by the National Research Foundation of Korea (NRF-2014R1A2A2A), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3414).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Son, SM., Ha, SY., Yoo, HY. et al. Prognostic impact of MYC protein expression in central nervous system diffuse large B-cell lymphoma: comparison with MYC rearrangement and MYC mRNA expression. Mod Pathol 30, 4–14 (2017). https://doi.org/10.1038/modpathol.2016.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.56
This article is cited by
-
MYC, BCL2, and BCL6 rearrangements in primary central nervous system lymphoma of large B cell type
Annals of Hematology (2019)
-
Novel synthetic 4-chlorobenzoyl berbamine inhibits c-Myc expression and induces apoptosis of diffuse large B cell lymphoma cells
Annals of Hematology (2018)