Abstract
Patients with clinically insignificant prostate cancer remain a major over-treated population. PTEN loss is one of the most recurrent alterations in prostate cancer associated with an aggressive phenotype, however, the occurrence of PTEN loss in insignificant prostate cancer has not been reported and its role in the separation of insignificant from significant prostate cancer is unclear. An integrated analysis of PTEN loss was, therefore, performed for structural variations, point mutations and protein expression in clinically insignificant (48 cases) and significant (76 cases) prostate cancers treated by radical prostatectomy. Whole-genome mate pair sequencing was performed on tumor cells isolated by laser capture microdissection to characterize PTEN structural alterations. Fluorescence in situ hybridization probes were constructed from the sequencing data to detect the spectrum of these PTEN alterations. PTEN loss by mate pair sequencing and fluorescence in situ hybridization occurred in 2% of insignificant, 13% of large volume Gleason score 6, and 46% of Gleason score 7 and higher cancers. In Gleason score 7 cancers with PTEN loss, PTEN alterations were detected in both Gleason pattern 3 and 4 in 57% of cases by mate pair sequencing, 75% by in situ hybridization and 86% by immunohistochemistry. PTEN loss by sequencing was strongly associated with TMPRSS2-ERG fusion, biochemical recurrence, PTEN loss by in situ hybridization and protein loss by immunohistochemistry. The complex nature of PTEN rearrangements was unveiled by sequencing, detailing the heterogeneous events leading to homozygous loss of PTEN. PTEN point mutation was present in 5% of clinically significant tumors and not in insignificant cancer or high-grade prostatic intraepithelial neoplasia. PTEN loss is infrequent in clinically insignificant prostate cancer, and is associated with higher grade tumors. Detection of PTEN loss in Gleason score 6 cancer in a needle biopsy specimen indicates a higher likelihood of clinically significant prostate cancer.
Similar content being viewed by others
Main
The tumor suppressor gene, PTEN (phosphatase and tensin homolog on chromosome 10), a phosphoinositide 3-phosphatase, negatively regulates the PI3K/AKT signaling pathway and functions as a tumor suppressor. It is one of the most commonly altered genes in prostate cancer. Mutations in PTEN occur in up to 10% of primary prostate cancers while PTEN deletion occurs in 10–70% of surgically treated cancers and over 50% of metastatic prostate cancers.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 PTEN deletion is associated with poorer cancer specific outcomes, and there is an association of deletion with increasing stage and Gleason score.7, 9, 12, 13, 14, 15, 16 Although associated with later stage and higher grade tumors, the frequency of PTEN loss is reported in 3–10% of Gleason score 6 cancers, and ~15% of confined pT2 tumors.12, 15 In regards to the proposed precursor lesion, high-grade prostatic intraepithelial neoplasia, the data are inconsistent and limited with five studies to date. In three studies that used fluorescence in situ hybridization for the evaluation of PTEN status, deletion was identified in 9, 13, and 23% cases of high-grade prostatic intraepithelial neoplasia.7, 10, 17 These findings in addition to mouse models that showed the development of high-grade prostatic intraepithelial neoplasia-like lesions following PTEN inactivation through deletion and mutation suggested that PTEN loss is an early event in prostate carcinogenesis, and potentially associated with more aggressive disease.18, 19, 20, 21 However, a study by McMenamin et al and by Lotan et al using immunohistochemistry indicate that in high-grade prostatic intraepithelial neoplasia, PTEN protein is intact suggesting that PTEN loss is a later event.22, 23
As PTEN loss is associated with aggressive prostate cancer, it could potentially have a role in separating patients that have significant prostate cancer from those with insignificant cancer. However, there remain important questions in regards to PTEN deletion and mutation and risk of prostate cancer significance and progression. Although PTEN deletion is reported in Gleason score 6 and confined prostate cancer, the frequency of PTEN mutation and deletion is unknown in insignificant prostate cancer (defined in radical prostatectomy specimens as tumors that are Gleason score 6, confined, and tumor volume <0.5 cm3). In addition, recent studies of contemporary Gleason grading and studies of Gleason score 6 cancer and outcome following surgery, suggest that studies have potentially overestimated the frequency of PTEN deletion in Gleason score 6 cancer by under-grading Gleason score 7 and higher grade cancers.24 Recently the presence of PTEN protein loss in Gleason score 6 needle biopsy specimens has been associated with increased likelihood of Gleason score upgrading in radical prostatectomy specimens.25 Understanding the relationship of PTEN loss, cancer significance and the relationship of Gleason pattern 3 and Gleason pattern 4 in regards to shared PTEN alterations in Gleason score 7 and higher cancer has important implications in the assessment of prostate needle biopsy specimens where only a small portion of a tumor is sampled.
The primary objectives of this study were to characterize the PTEN status in insignificant and significant prostate cancer from radical prostatectomy specimens using whole-genome mate pair sequencing, Sanger sequencing, fluorescence in situ hybridization and immunohistochemistry to determine the frequency of PTEN abnormalities in insignificant and significant prostate cancer including structural alterations and mutations. The additional objective was to understand the relationship of Gleason pattern 3 and Gleason pattern 4 in Gleason score 7 cancers in regard to PTEN loss to determine if understanding PTEN status could have a role in determining prostate cancer significance and stratifying risk of progression in newly diagnosed prostate cancer patients.
Materials and methods
Tissue Collection and Processing
Consecutive cases of insignificant Gleason score 6 (48 cases), large volume Gleason score 6 (23 cases), Gleason score 7 (29 cases), and Gleason score 8 and higher cancers (25 cases) were selected from the Mayo Clinic prostate cancer frozen repository. Insignificant prostate cancer was defined as Gleason score 6, stage pT2 and tumor volume <0.5 cm3. Processing of radical prostatectomy specimens occurred as previously described.26 All specimens were handled in the fresh state and analyzed initially with a frozen microtome technique. The prostate was inked, serially sectioned in the horizontal plane, and right apex, left apex and bladder base margins (perpendicular) were examined microscopically followed by standard sections from the inferior, mid and superior posterior prostate and right and left vesicles. In cases of insignificant Gleason score 6 cancer, the entire posterior prostate was examined with additional sections of the right and left anterior prostate. The freezing microtome technique conserves tissues such that a top and bottom created from cutting allows for the collection of fresh insignificant prostate cancer (unfrozen top section collected for analysis). For large volume Gleason score 6, Gleason score 7 and Gleason score 8 and higher tumors, the dominant index tumor in the gland was collected. The tops and bottoms of the tissue cut by the microtome technique were processed to formalin-fixed paraffin-embedded tissue sections for review after routine processing.
Isolation of DNA and Mate Pair Sequencing
Tumor was collected by laser capture microdissection (Arcturus instrument) from 10 μm unstained fresh-frozen sections and DNA amplified directly from cells, as previously described.27, 28, 29, 30 In Gleason score 7, the Gleason pattern 3 and Gleason pattern 4 were collected and analyzed separately.28 Libraries were prepared using the Illumina Mate Pair (MP) Kit following the manufacturer’s instructions and sequenced as two libraries per lane on the Illumina HiSeq platform.27, 28, 29, 30, 31 Data were processed using previously described binary indexing mapping algorithms developed in our group.32 The read-to-reference-genome-mapping algorithm was modified to map both mate pair sequencing reads across the whole genome. Discordant mapping mate pair sequencing reads that were covered by at least five associates were identified for further analysis. Concordant mapping mate pair sequencing reads were used to determine frequency coverage levels across the genome.31, 32
Fluorescence In Situ Hybridization
The diverse repertoire of PTEN structural variants detected by whole-genome mate pair sequencing was used to develop a unique PTEN fluorescence in situ hybridization probe set that would detect as many structural variants as possible in the corresponding formalin-fixed paraffin-embedded prostate cancer. Probes were designed and prepared in a similar manner to probes developed in our laboratory. Briefly, bacterial artificial chromosomes corresponding to the regions of interest overlying PTEN and adjacent regions were obtained from ResGen Invitrogen. DNA was isolated using the QIAGEN Plasmid Maxi Kit and labeled using the Vysis Nick Translation kit and either SpectrumGreen-dUTP or SpectrumOrange-dUTP (Vysis). Each BAC clone was tested individually by hybridization to normal metaphases. Polymerase chain reaction confirmed clones covered the regions of interest. Centromere 10 status was assessed using commercially available probe (CEP10, Abbott Molecular). Tissue sections (5 μm) were deparaffinized at 90 °C, immersed twice in xylene, twice in 100% ethanol, dried, microwaved in 10 mmol/l citric acid (10 min), pretreated with 2x saline-sodium citrate buffer (5 min), digested (40 min) with DigestALL (Invitrogen) and washed. Slides were denatured at 80 °C (5 min) and hybridized at 37 °C overnight in humidified incubator. DAPI1 (10%; Abbott) was used for counter staining. Before hybridization, tumor was identified and labeled by a pathologist on H&E slides that corresponded with sections used for in situ hybridization. Each case was scored by two technicians in a blinded manner. Before scoring, technicians first scanned the entire region of interest to identify potential PTEN status heterogeneity within the tumor. Then the cases were scored with each technician scoring 100 interphase nuclei from representative areas. Each cell was scored for all probes simultaneously. Scoring was done blinded to whole-genome mate pair sequencing, Sanger sequencing, and immunohistochemical results. In Gleason score 7 cancer, Gleason pattern 3 and Gleason pattern 4 were scored separately.
Immunohistochemistry
Formalin-fixed paraffin-embedded 5 μm sections from the same tissue block used for in situ hybridization were incubated in EDTA-containing antigen retrieval buffer (32 min), followed by application of primary rabbit monoclonal antibody (PTEN clone D4.3; Cell Signaling) at 1:200 dilution made with Dako Background Reducing Diluent for (32 min) at 37 °C. Immunostaining was performed on the Ventana BenchMark XT Stainer with Ventana Optiview Plus Amp detection system followed by Ventana DAB, hematoxylin (8 min) and bluing agent (4 min). Cancer was scored independently by two pathologists (JCC and EPC) as the percentage of cells showing markedly decreased or absent PTEN staining, and the tumor was considered to show PTEN loss by immunohistochemistry if any area (or percentage of) tumor lacked PTEN staining. Scoring was done blinded to whole-genome mate pair sequencing, fluorescence in situ hybridization and Sanger sequencing results. In Gleason score 7, the Gleason pattern 3 and Gleason pattern 4 were scored separately.
Sanger Sequencing
Tumor and associated normal prostate DNA (10 ng) purified from the laser capture microdissected fresh-frozen tissue was used to assess PTEN gene mutation status. Bi-directional Sanger sequence analysis was performed to test for the presence of a mutation in all coding regions and intron/exon boundaries of the PTEN gene (GenBank accession number NM_000314; NG_007466.2).
Results
Clinical and Pathological Features
Clinico-pathologic features of the patients were available for 110 (87%) of the cases (Table 1). As expected preoperative serum PSA and pathological tumor stage correlated with Gleason score. The mean tumor size for the large volume Gleason score 6 was similar to that of the Gleason score 7 group at 3.9 and 3.7 cm3, respectively, whereas the Gleason score 8 and higher tumors had a significantly greater mean volume at 10.4 cm3. By definition, all insignificant Gleason score 6 cancers were <0.5 cm3. Clinical outcome data demonstrated that biochemical recurrence and systemic progression correlated with Gleason score (Table 1). Three (3%) patients died of prostate cancer, all from the Gleason score 8 and higher group. Survival curves with biochemical recurrence as an end point exemplify the stratification of insignificant and large volume Gleason score 6 from higher Gleason score cancers (Figure 1).
Survival curves with biochemical recurrence as an end point exemplifying the stratification of insignificant Gleason 6 and large volume Gleason score 6 from higher Gleason score cancers. Percentage of patients free of biochemical recurrence (y-axis) plotted against years from surgery (x-axis) for four patient groups; patients with insignificant tumor (solid gray line), all Gleason score 6 tumors (solid black line), large volume Gleason score 6 together with Gleason score 7+ tumors (dashed gray line) and Gleason score 7 and higher tumors (dashed black line).
Large Genomic Rearrangements of PTEN
Whole-genome mate pair sequencing provided in-depth profiling of large genomic rearrangements potentially affecting PTEN function in the 126 prostate cancers. In total, 29 (23%) cases showed genomic alterations that predicted loss of function of PTEN. A detailed description of genomic breakpoints affecting chromosome 10 and the PTEN locus is provided in Table 2 and depicted in Figure 2. Polymerase chain reaction validations on selected PTEN deletions confirmed the validity of detected events (Supplementary Figure 1). The majority of cases with PTEN abnormalities (92%) fell in the Gleason score 7 and 8 and higher groups (14 (48%) and 11 (44%) cases, respectively). Just a single insignificant Gleason score 6 cancer (2%) and three large volume Gleason score 6 (13%) cancers had PTEN loss. Overall, events were more common in higher Gleason grade tumors, detected in 11 (11%) of 101 Gleason pattern 3 cancers and 25 (47%) of 53 Gleason patterns 4/5 cancers.
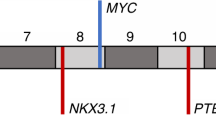
Rearrangements within the 10q23 locus. (a) Repertoire of rearrangements observed in PTEN in the prostate tumors studied from whole-genome mate pair sequencing. PTEN region is presented spanning from 84 to 98 Mb of 10q23 with the position of PTEN indicated. Rearrangements for specific tumors listed are presented as deletions (solid lines), inversions (dashed lines), or spans of collective complex events (arrowed triple lines). Specific intrachromosomal breakpoints across 10q23 are marked by circles and numbers indicate translocation to different chromosome. Arrowed lines indicate additional events outside the 10q23 region presented. Cases are presented with alternative gray and black coloring for clarity. In the upper Gleason score 7 group, a and b refer to the Gleason pattern 3 and Gleason pattern 4 components, respectively. (b) Positioning of PTEN probes designed for fluorescence in situ hybridization.
Mate pair reads spanning PTEN and predicting deletions were detected in 26 of the 29 cases (Table 2). PTEN deletion events ranged from single deletion events spanning the PTEN gene, to more complex rearrangements, with multiple rearrangements across the chromosome 10q23 region. In many cases multiple breakpoint events occurred surrounding PTEN, predicting more complex rearrangement events leading to loss of PTEN, with these more complex events more commonly present in Gleason pattern 4 than Gleason pattern 3. In addition to intrachromosomal rearrangements, interchromosomal translocations occurred adjacent to PTEN, often in association with intrachromosomal events. Multiple potentially independent events within a single tumor indicated homozygous loss of PTEN. Genomic breakpoints hitting directly within the PTEN gene-coding region were observed in 16 tumors, 13 involving rearrangements within chromosome 10 and 4 by interchromosomal translocations fusing the PTEN coding region to different chromosomes. Although the effect of these alterations on PTEN function is sometimes difficult to interpret, copy number variation levels from the mate pair sequencing data confirmed the loss of the PTEN. Homozygous loss of PTEN was predicted in nine tumors, all Gleason score 7 and higher. Chromothripsis or chromosomal catastrophe is a single catastrophic event resulting in multiple genomic rearrangements, and associated with poor outcome in a number of cancers. When defined by the following: affected locus >10 megabases, number of clustered breakpoints >12, breakpoints involving one or two chromosomes, and inclusion of alternating copy number states, insertions and loss of heterozygosity, none of the cases showed involvement of the 10q23 region as we have previously reported.29
In Gleason score 7 tumor with PTEN loss (14 cases), all Gleason pattern 4 had rearrangements involving PTEN, compared with 8 (57%) of the corresponding Gleason pattern 3. In these eight cases, 5 (63%) shared identical PTEN events within Gleason pattern 3 and Gleason pattern 4, whereas three cases contained unique events in the Gleason pattern 3 and the Gleason pattern 4. Unique events in PTEN were often seen in addition to shared lineage events, indicative of tumor progression. This observation was also evident in coverage frequency plots across PTEN, with additional deletion events in Gleason pattern 4 in the background of events in the Gleason pattern 3, indicative of progressive bi-allelic loss of PTEN (Supplementary Figure 2).
In addition to evaluation of PTEN status in the tumor, whole-genome mate pair sequencing was performed on high-grade prostatic intraepithelial neoplasia collected by laser capture microdissection. High-grade prostatic intraepithelial neoplasia was collected from 30 patients consisting of 15 insignificant Gleason score 6, six large volume Gleason score 6, and nine Gleason score 7 and higher cases. High-grade prostatic intraepithelial neoplasia was collected in the same section as tumor but where it was clearly delineated from the cancer to allow collection by laser capture microdissection without contamination by tumor. Rearrangements resulting in PTEN loss were not observed in any cases of high-grade prostatic intraepithelial neoplasia.
Heterogeneity of PTEN Loss and Correlation with TMPRSS2-ERG Fusion
TMPRSS2-ERG fusion was present in ~50% of cases from the mate pair sequencing data (Table 2). Although not predictive of tumor grade, TMPRSS2-ERG fusion correlated with PTEN loss (Figure 3a). Specifically in Gleason score 6 and Gleason score 7 cancer, nearly 80% of cases with loss of PTEN, manifested a TMPRSS2-ERG fusion. However, correlation was lower in the Gleason score 8 and higher group where just a 45% correlation was observed, consistent with previous reports.15
Detection of PTEN loss by mate pair sequencing, fluorescence in situ hybridization (FISH) and immunohistochemistry. Percentage of cases in the insignificant Gleason score 6, large volume Gleason score 6, Gleason score 7, Gleason score 8 and higher or all tumor groups associated with: (a) TMPRSS2-ERG Fusion (ERG positive) and PTEN loss by mate pair sequencing. Cases with TMPRSS2-ERG fusions (light gray bars), PTEN loss (black bars) or both (dark gray bars). (b) PTEN loss by mate pair sequencing (black bars), FISH (light gray bars) or both (dark gray bars). (c) PTEN loss by mate pair sequencing (black bars), immunohistochemistry (light gray bars) or both (dark gray bars). (d) PTEN loss by mate pair sequencing (black bars), FISH (light gray bars), immunohistochemistry (dark gray bars), or shared loss by two (hatched bar) or three (dotted bar) techniques. (e and f) Comparison of PTEN loss by MP, FISH and immunohistochemistry in cases where all three techniques applied as measures by any loss (e) or as single and double copy losses (f).
Consistent with previously reported data from our group, identical TMPRSS2-ERG fusions were present in both Gleason pattern 3 and Gleason pattern 4 in 11 of the 14 Gleason score 7 cases29 (Table 2). An overall assessment of lineage between the Gleason pattern 3 and Gleason pattern 4 in Gleason score 7 cancer utilizing all shared rearrangements enabled assignment of high, medium, low or no lineage calls (Table 2). Overall, unique PTEN events in Gleason pattern 3 and Gleason pattern 4 correlated with lineage. For example, in cases PR23, PR31, and PR62, where no shared PTEN events were observed between the Gleason patterns in a tumor, the lineage between these patterns was assessed low or none. Conversely, all high/medium lineage cases presented with either shared identical PTEN events or no events in Gleason pattern 3 and Gleason pattern 4.
PTEN Point Mutations
To investigate the involvement of point mutations in the PTEN gene, each exon and the promoter element of every tumor, high-grade prostatic intraepithelial neoplasia and associated normal prostate tissue of the 126 cases were screened by bi-directional Sanger sequencing. Six (5%) cases possessed mutations in the PTEN gene (Table 2 and Table 3), five Gleason score 7 tumors and one Gleason score 9 tumor. Just one of the mutations in the Gleason score 7 group was present in the Gleason pattern 3 component, and in the remaining four; the point mutation was in Gleason pattern 4 only. All of the cases with PTEN point mutation presented with additional PTEN loss by mate pair sequencing (Table 2) indicating a mechanism of homozygous loss of PTEN in 5% of cases. No mutations were observed in high-grade prostatic intraepithelial neoplasia or normal tissues.
Evaluation of PTEN Rearrangements by Fluorescence In Situ Hybridization
Guided by the array of structural alterations identified by whole-genome mate pair sequencing (Figure 2), a fluorescence in situ hybridization probe set was developed to validate the PTEN rearrangements and performed on formalin-fixed paraffin-embedded tissues from 97 of the 126 cases. The PTEN in situ hybridization results showed high concordance with the mate pair sequencing results (Figure 3b). Twenty-two of the 29 cases with PTEN deletion by mate pair sequencing were evaluated by fluorescence in situ hybridization with equivalent Gleason grades observed in the formalin-fixed paraffin-embedded tissues (Table 4). Of these 20 (91%) showed PTEN alterations by both in situ hybridization and mate pair sequencing. The two cases that did not correlate were both from Gleason score 7 group (Supplementary Table 1). In the remaining cases where no PTEN deletion was detected by mate pair sequencing, 10 (10%) showed PTEN loss by fluorescence in situ hybridization (Table 4) with the majority in the Gleason score 6 groups (Figure 3b). In addition, in Gleason score 7 tumors with PTEN loss, Gleason pattern 3 and Gleason pattern 4 shared loss in 75% of cases. This is the result of limited sampling of the laser capture microdissected samples compared with scanning the entire tumor in fluorescence in situ hybridization resulting in the identification of subpopulations within Gleason pattern 3 that show PTEN loss.
Immunohistochemical Staining for PTEN
Immunohistochemical staining for PTEN was additionally performed on the formalin-fixed paraffin-embedded tissues from 107 (85%) of the 126 cases studied by mate pair sequencing. The tumor was considered to show loss of PTEN if any tumor showed markedly decreased or absent staining. Loss of PTEN staining was observed in 35 (33%) cases (Figure 3c and Table 4) with the majority of tumors (95%) showing >10% loss. In regards to staining heterogeneity, six (17%) cases showed <10% staining loss, six (17)% showed 10–50% staining loss, and 23 (66%) showed >50% staining loss. Of the 29 cases with PTEN loss by mate pair sequencing, 26 were assessed by immunohistochemistry, with 23 (89%) confirming a loss of PTEN expression (Figure 3c). The three cases where no loss of PTEN expression was observed by mate pair sequencing consisted of one case each from the insignificant Gleason score 6, large volume Gleason score 6, and Gleason score 7 groups (Supplementary Table 1). The insignificant Gleason score 6 case (PR65) also showed intact PTEN by fluorescence in situ hybridization. Of the remaining 82 cases surveyed, 12 (11%) showed loss of PTEN expression in the absence of detected mate pair sequencing rearrangements. From these 12 cases, 10 were also analyzed by fluorescence in situ hybridization, with five showing loss of PTEN. In addition, by immunohistochemistry, 86% of cases with PTEN loss in the Gleason pattern 4 also observed loss in the adjacent Gleason pattern 3. No PTEN expression loss was observed in high-grade prostatic intraepithelial neoplasia.
Concordance of PTEN Loss by Mate Pair Sequencing, Fluorescence In Situ Hybridization, and Immunohistochemistry
The concordance in detection of PTEN loss between each technique was high, ranging from 85.4 to 86.6% (Table 4). Concordance between the three techniques was higher in the higher grade (Gleason score 7 and higher) tumors, with less concordance in Gleason score 6 (Figure 3d) with additional cases of PTEN loss detected by both fluorescence in situ hybridization and immunohistochemistry that were not detected by mate pair sequencing. Within the 96 cases assayed by all three techniques, 23 (23%) cases showed PTEN alterations by mate pair sequencing, compared with 29 (30%) and 29 (30%), by fluorescence in situ hybridization and immunohistochemistry, respectively. In the cases where PTEN loss was detected by mate pair sequencing confirmatory loss by both fluorescence in situ hybridization and immunohistochemistry was 100%. The additional cases detected by immunohistochemistry and fluorescence in situ hybridization were predominantly in the insignificant Gleason score 6 and large volume Gleason score 6 groups (Figure 3d). Of the 15 cases that showed PTEN alterations by fluorescence in situ hybridization or immunohistochemistry not detected by mate pair sequencing, ten were Gleason score 6 and just five showed loss by both immunohistochemistry and fluorescence in situ hybridization. The status of each case determined by each technique is presented in Figure 3e along with hemizygous or homozygous loss of PTEN.
PTEN Loss and Biochemical Recurrence
Survival curves contrasting PTEN deletion detected by mate pair sequencing with biochemical recurrence as an end point demonstrated significance correlation (Figure 4a). Nearly 60% of patients with PTEN deletions by mate pair sequencing had a biochemical recurrence, compared with ~25% of patients without a deletion. In patients with Gleason score 7 and higher tumors, PTEN loss remained predictive of biochemical recurrence with 80% of patients whose tumors contained PTEN loss developing biochemical recurrence compared with 55% of patients whose tumor had intact PTEN (Figure 4b). There was no significant difference in the association of PTEN loss and biochemical recurrence by the three techniques (Supplementary Figure 3). The significance of TMPRSS2-ERG fusion status was also investigated (Figure 4c). Although the sample is small, a worse prognosis occurred in patients with that had a TMPRSS2-ERG fusion and loss of PTEN.
Survival curves of association of biochemical recurrence with PTEN loss. (a) PTEN loss from mate pair sequencing and biochemical recurrence in all cases. Case grouped according to PTEN normal (gray) or loss (black). (b) PTEN loss from mate pair sequencing data and biochemical recurrence in Gleason score 7 and above cases. Case grouped according to PTEN normal (gray) or loss (black). (c) Stratification of PTEN loss and TMPRSS2-ERG fusion (ERG) with biochemical recurrence. Case grouped according to PTEN loss/ERG positive (black solid) PTEN loss/ERG negative (black hashed) PTEN normal/ERG positive (gray solid) PTEN normal/ERG negative (gray hashed).
Discussion
Loss of the tumor suppressor, PTEN, has long been associated with high-grade prostate cancer. However, little is known about the frequency and impact of PTEN deletions and mutation in clinically insignificant prostate cancer. In this study we used a whole-genome sequencing approach to detect the genomic breakpoints in 126 prostate cancers treated by radical prostatectomy consisting of insignificant and large volume Gleason score 6, Gleason score 7 and 8 and higher grade cancers and compared sequencing results to fluorescence in situ hybridization and immunohistochemistry. The whole-genome mate pair sequencing approach enabled detection of all large genomic rearrangements impacting PTEN, and provided data to construct fluorescence in situ hybridization probes to cover the majority of alterations. The results demonstrate that PTEN loss by mate pair sequencing, and fluorescence in situ hybridization and protein loss by immunohistochemistry is low in clinically insignificant prostate cancer, and loss (including homozygous loss) increases with increasing Gleason score. In addition, we did not identify any PTEN point mutations in clinically insignificant prostate cancer, and loss occurred in higher grade tumors associated with PTEN structural alterations resulting in homozygous PTEN loss. Finally, in Gleason score 7 cancer with PTEN alterations, the PTEN loss was detected in both Gleason pattern 3 and Gleason pattern 4 in 57% of cases by mate pair sequencing, 75% by fluorescence in situ hybridization, and 86% by immunohistochemistry.
In contrast to the well-characterized TMPRSS2-ERG fusion, our data indicates that rearrangements resulting in PTEN loss are highly heterogeneous between and within tumors (Table 2, Figure 2 and Supplementary Figure 2). Rearrangements were observed that spanned PTEN, or occurred within the 10q23 locus adjacent to or directly within the PTEN gene, with minimal similarity between cases. In addition to rearrangements solely within chromosome 10, many interchromosomal rearrangements were observed adjacent to or directly within PTEN, although no commonality was seen in partnering chromosomes between tumors. Complex events involving both multiple intra- and inter-chromosome 10 rearrangement events were also common resulting in genomic restructuring that was difficult to interpret but copy number variation analysis of mate pair sequencing data achieved by using frequency read-coverage from concordantly mapping mate pairs, confirmed loss of PTEN. Homozygous PTEN loss was also frequent and more common in the Gleason score 8 and higher tumors. Progressive losses were often observed in the Gleason score 7 group, where shared single loss events were evident between the Gleason pattern 3 and Gleason pattern 4, but with additional progressive losses within Gleason pattern 4 (Supplementary Figure 2 and Table 2). In no case, did we observe that chromothripsis or chromoplexis was a mechanism of PTEN loss.
PTEN loss was detected in 23% of cases by mate pair sequencing, and 31% of cases by fluorescence in situ hybridization and protein loss in 33% of cases by immunohistochemistry. In clinically insignificant cancer, loss was detected in one, four and two cases by mate pair sequencing, fluorescence in situ hybridization and IHC, respectively. However, only one of the four cases detected by fluorescence in situ hybridization demonstrated a loss of PTEN expression in the immunohistochemistry. Interestingly, the clinically insignificant cancer detected for PTEN loss by mate pair sequencing, was also detected by in situ hybridization, but no loss of PTEN expression was observed by immunohistochemistry. The higher detection of PTEN alterations by in situ hybridization and loss of protein by immunohistochemistry can be explained by differences in the three techniques. Mate pair sequencing used small samples of pure population of tumor cells obtained by laser capture microdissection of frozen tissues compared with whole-section visual analysis by in situ hybridization and immunohistochemistry. Assessment of whole sections allowed for the detection of populations of tumor cells that had PTEN loss that were likely missed by laser capture microdissection. However, the three techniques showed high concordance with infrequent loss of PTEN and protein in insignificant cancer, and a significant association with higher Gleason score and patient outcome. Comparison with other studies is difficult because of technical differences. In our study, we used whole tumor sections rather than tissue microarrays, and developed specific probes based on the whole-genome mate pair sequencing data. Studies have shown a concordance of fluorescence in situ hybridization and immunohistochemistry for PTEN and protein loss but lower than our study. For instance, Lotan et al found that 45% of tumors that showed PTEN loss by immunohistochemistry had intact PTEN by fluorescence in situ hybridization.33 This discordance can be explained by the complexity of the PTEN alterations. The heterogeneity of PTEN alterations in prostate cancer was appreciated by Reid et al who developed a probe strategy similar to our own with probes overlying the 3′ and 5′ as well as a third probe overlying the PTEN gene.34 We confirm their hypothesis that gross rearrangements exist that cause disruption of PTEN in the absence of genomic deletion, and that a probe strategy that encompasses these larger rearrangements shows a higher concordance with immunohistochemistry, and explains the absence of PTEN protein in the setting of an intact PTEN as assessed by a single break-apart fluorescence in situ hybridization probe. Interestingly, in our study, both fluorescence in situ hybridization and immunohistochemistry demonstrated that in Gleason score 7 cancer the Gleason pattern 3 and 4 shared PTEN alterations and loss of protein in 75 and 86% of cases, respectively. This finding has significant implications in the interpretation of PTEN status in needle biopsy specimens. Recently, Lotan et al compared patients with Gleason score 6 on the needle biopsy specimen that had an upgrading of Gleason score in the radical prostatectomy specimen to patients that had Gleason score 6 in the needle biopsy and radical prostatectomy specimen.23 They found that PTEN protein loss in the needle biopsy specimen was significantly associated with upgrading in the radical prostatectomy specimen. This is an important finding particularly in patients who are candidates of active surveillance where detection of a PTEN alteration on needle biopsy likely indicates the presence of clinically significant prostate cancer. One important limitation in assessing PTEN abnormalities in needle biopsy specimens includes the heterogeneity of PTEN loss in prostate cancers, and the limited sampling by biopsy. Indeed, although we did not specifically address the heterogeneity of PTEN alterations by fluorescence in situ hybridization, we did see heterogeneity in staining by immunohistochemistry. Although, nearly two-thirds of cases showed >50% of tumor staining loss, a third showed <50% staining loss. This heterogeneity will certainly impact the detection of PTEN alterations and lead to a significant false negative rate. However, we believe the detection of a PTEN alteration will remain clinically useful, and indicate the likelihood of clinically significant prostate cancer. We are currently applying our PTEN fluorescence in situ hybridization probe set to a large cohort of matched needle biopsy and radical prostatectomy specimens to address the issues of heterogeneity, and sensitivity and false negative rates.
To the best of our knowledge, this is the first study to assess PTEN point mutations in clinically insignificant prostate cancer and high-grade prostatic intraepithelial neoplasia, and mutations were not identified in either. Although a less common pathway for PTEN loss, point mutations in the PTEN gene is important as a pathway for homozygous loss of PTEN. Six of 126 cases contained deleterious mutations, all in the Gleason score 7 and higher cancers. Mutations were generally late, occurring predominantly in the Gleason pattern 4 and 5. In contrast to PTEN structural alterations, only one case of Gleason score 7 shared a point mutation within Gleason pattern 3 and 4. In addition, these PTEN mutations all occurred in cases with additional PTEN deletions indicating a mechanism of achieving homozygous loss of PTEN function. Although not investigated in this study, epigenetic silencing of PTEN has been described as a further mechanism of PTEN expression loss, which could also function in association with mutations to drive homozygous PTEN loss.6, 35 Haploinsufficiency of PTEN has been extensively associated with aggressive prostate cancer phenotype.9, 10, 17, 36, 37, 38, 39, 40 Cells with haploinsufficiency of PTEN have been reported to be more unstable, defining a role for PTEN in maintaining chromosomal stability, as well increasing PI3K/AKT upregulation.36, 37, 38, 39, 40, 41
Although PTEN loss in prostate cancer has been reported as a progressive later event in prostate cancer, contradictory literature exists on the occurrence in high-grade prostatic intraepithelial neoplasia.10, 17, 23 In this study PTEN loss was assessed in 30 patients with high-grade prostatic intraepithelial neoplasia adjacent to the prostate cancer by whole-genome mate pair sequencing, Sanger sequencing, fluorescence in situ hybridization and immunohistochemistry. We identified no loss of PTEN expression, or any PTEN structural variations or point mutation in the high-grade prostatic intraepithelial neoplasia. This confirms recent reports, and suggests that PTEN alterations reported in high-grade prostatic intraepithelial neoplasia may be occurring in intraductal carcinoma.23
In summary, whole-genome mate pair sequencing identified the heterogeneous and complex rearrangements resulting in PTEN loss in prostate cancer across the disease spectrum. PTEN deletion by whole-genome mate pair sequencing, fluorescence in situ hybridization and PTEN immunohistochemistry is infrequent in clinically insignificant prostate cancer and significantly correlated with increasing Gleason score and biochemical recurrence. Correlation between the three techniques was high. Fluorescence in situ hybridization and immunohistochemistry detected additional cases with PTEN loss, indicating a higher false negative rate by the whole-genome technique due to sampling. In addition, using an in situ hybridization probe strategy that encompasses the spectrum of large chromosomal alterations detects higher frequency of tumors with PTEN deletions. Currently in our practice, we have implemented both the PTEN fluorescence in situ hybridization assay and immunohistochemistry. The PTEN in situ hybridization assay is performed in a cytogenetics laboratory with cytotechnologists dedicated to the assessment of clinical in situ hybridization assays. The in situ hybridization assay uses breakapart probes, and we believe can be readily implemented in clinical laboratories routinely performing clinical assays. It is important to recognize that small deletions can result in a diminished but not completely absent probe signal. We are currently in the process of comparing both assays on needle biopsy specimens. Whole-genome mate pair sequencing requires fresh-frozen tissue and is not applicable to biopsy specimens. Finally, detection of PTEN loss in Gleason score 6 in a needle biopsy specimen indicates a higher likelihood of clinically significant prostate cancer.
Accession codes
References
Cairns P, Okami K, Halachmi S et al. Frequent inactivation of PTEN/MMAC in primary prostate cancer. Cancer Res 1997;57:4997–5000.
Feilotter HE, Nagai MA, Boag AH et al. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene 1998;16:1743–1782.
Pesche S, Latil A, Muzeau F et al. PTEN/MMAC/TEP1 involvement in primary prostate cancers. Oncogene 1998;16:2879–2883.
Suzuki H, Freije D, Nusskern DR et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple prostate cancer tissues. Cancer Res 1998;58:204–209.
Wang SI, Parsons R, Ittmann M . Homozygous deletion of PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res 1998;4:811–815.
Whang YE, Wu X, Suzuki H et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA 1998;95:5246–5250.
Han B, Mehra R, Lonigro RJ et al. Fluorescence in situ hybridization study shows association of PTEN deletion and ERG rearrangement during prostate cancer progression. Mod Pathol 2009;22:1083–1093.
Taylor BS, Schultz N, Hieronymus H et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22.
Yoshimoto M, Cunha IW, Coudry RA et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 2007;97:678–685.
Yoshimoto M, Cutz JC, Nuin PA et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 2006;169:128–137.
Suzuki H, Freije D, Nusskern DR et al. Interfocal heterogeneity of PTEN/MMAC gene alterations in multiple metastatic prostate cancer tissues. Cancer Res 1998;58:204–209.
Krohn A, Diedler T, Burkhardt L et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol 2012;181:401–412.
McCall P, Witton CJ, Grimsley S et al. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer 2008;99:1296–1301.
Sircar K, Yoshimoto M, Monzon FA et al. PTEN genomic deletion is associated with p-AKT and AR signaling in poorer outcome, hormone refractory prostate cancer. J Pathol 2009;218:678–684.
Yoshimoto M, Joshua AM, Cunha IW et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21 (12):1451–1460.
Reid AH, Attard G, Ambroisine L et al. Molecular characterization of ERG ETV1, and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer 2010;102:678–684.
Bismar TA, Yoshimoto M, Vollmer R et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Intl 2010;107:4077–4485.
Di Cristofano A, Pesce B, Cordon-Cardo C et al. PTEN is essential for embryonic development and tumor suppression. Nat Genet 1998;19:348–355.
Di Cristofano A, De Acetis M, Koff A et al. PTEN and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nature Gen 2001;27:222–224.
Kwabi-Addo B, Giri D, Schmidt K et al. Haploinsufficiency of the PTEN tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci USA 2001;98:11563–11568.
Luchman HA, Benediktsson H, Villemaire AC et al. The pace of prostatic intraepithelial neoplasia development is determined by the timing of PTEN tumor suppressor gene excision. PloS One 2008;3:3940.
McMenamin ME, Soung P, Perera S et al. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res 1999;59:4291–4296.
Lotan TL, Gumuskaya B, Rahimi H et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol 2012;26:587–603.
Ross HM, Kryvenko ON, Cowan JE et al. Do adenocarcinomas of the prostate with Gleason score (GS) ≤6 have the potential to metastasize to lymph nodes? Am J Surg Pathol 2012;36:1346–1152.
Lotan TL, Carvalho FL, Peskoe SB et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2015;28:128–137.
Sebo TJ, Cheville JC, Riehle DL et al. Perineural invasion and MIB-1 positivity in addition to Gleason score are significant preoperative predictors of progression after radical retropubic prostatectomy for prostate cancer. Am J Surg Pathol 2002;26:7–14.
Murphy SJ, Cheville JC, Zarei S et al. Mate pair sequencing of whole-genome-amplified DNA following laser capture microdissection of prostate cancer. DNA Res 2012;19:395–406.
Kovtun IV, Cheville JC, Murphy SJ et al. Lineage relationship of Gleason patterns in Gleason score 7 prostate cancer. Cancer Res 2013;73:3275–3284.
Kovtun IR, Murphy SJ, Johnson SH et al. Chromosomal catastrophe is a frequent event in clinically insignificant prostate cancer. Oncotarget 2015;6:29087–29096.
Murphy SJ, Hart SN, Lima JF et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 2013;145:1098–1109.
Vasmatzis G, Johnson SH, Knudson RA et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood 2012;120:2280–2289.
Drucker TM, Johnson SH, Murphy SJ et al. BIMA V3: an aligner customized for mate pair library sequencing. Bioinformatics 2014;30:1627–1629.
Lotan TL, Gurei B, Sutcliffe S et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 2013;17:6563–6573.
Reid AHM, Attard G, Brewer D et al. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol 2012;25:902–910.
Konishi N, Nakamura M, Kishi M et al. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol 2002;160:1207–1214.
Shen MM, Abate-Shen C . Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010;24:1967–2000.
Carver BS, Tran J, Gopalan A et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 2009;41:619–624.
Alimonti A, Carracedo A, Clohessy JG et al. Subtle variations in PTEN dose determine cancer susceptibility. Nat Genet 2010;42:454–458.
Squire JA . TMPRSS2-ERG and PTEN loss in prostate cancer. Nat Genet 2009;41:509–510.
Yoshimoto M, Ding K, Sweet JM et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol 2012;26:435–447.
Shah RB, Bentley J, Jeffery Z et al. Heterogeneity of PTEN and ERG expression in prostate cancer on core needle biopsies: implications for cancer risk stratification and biomarker sampling. Hum Pathol. 2015;46:698–706.
Acknowledgements
This publication was supported by funding from the Mayo Clinic Center for Individualized Medicine. We would also like to acknowledge the contributions of Bruce Eckloff and Robert Sikkink of the Mayo Medical Genomics Facility for their assistance with the Mate Pair sequencing.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Murphy, S., Karnes, R., Kosari, F. et al. Integrated analysis of the genomic instability of PTEN in clinically insignificant and significant prostate cancer. Mod Pathol 29, 143–156 (2016). https://doi.org/10.1038/modpathol.2015.136
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2015.136
This article is cited by
-
Hyperphosphorylated PTEN exerts oncogenic properties
Nature Communications (2023)
-
Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients
Journal of Translational Medicine (2020)
-
Merging new-age biomarkers and nanodiagnostics for precision prostate cancer management
Nature Reviews Urology (2019)
-
Distinct subtypes of genomic PTEN deletion size influence the landscape of aneuploidy and outcome in prostate cancer
Molecular Cytogenetics (2018)
-
Clinical implications of PTEN loss in prostate cancer
Nature Reviews Urology (2018)