Abstract
Liposarcoma represents a unique model insofar as some well-differentiated liposarcomas progress to non-lipogenic, so-called ‘dedifferentiated,’ forms. The well-differentiated and dedifferentiated family of liposarcomas demonstrates amplification of the chromosome subregion 12q13–q15 with resultant amplification of the MDM2 and CDK4 genes. However, the specific genetic changes that distinguish between well-differentiated and dedifferentiated liposarcomas are less well understood. To study the genetic changes in dedifferentiated liposarcomas, paired well-differentiated and dedifferentiated components of 29 tumors were analyzed separately by array-based comparative genomic hybridization. A bacterial artificial chromosome array at ∼1-Mb resolution was used. The genetic changes were compared with clinical presentation, grade of the dedifferentiated component and overexpression of MDM2 and CDK4. Most tumors (n=21, 72%) were retroperitoneal, with both components present at initial diagnosis (n=25, 86%). Eight tumors (28%) were classified as low-grade dedifferentiation. In four cases (14%), a well-differentiated liposarcoma preceded the presentation of the dedifferentiated tumor by 1–5 years. 12q13–q15 was amplified in all tumors. Using unsupervised hierarchical clustering of copy-number changes, all but two tumors showed close similarities between well-differentiated and dedifferentiated components, and segregated as pairs. Dedifferentiated components had more total amplifications (P=0.008) and a trend for gain at 19q13.2, but no genetic changes were significant in distinguishing between the two components. High-level amplifications of 1p21–32 (n=7, 24%), 1q21–23 (n=9, 31%), 6q23–24 (n=6, 21%) and 12q24 (n=3, 10%) were common, but none significantly correlated with differentiation. Presentation and grade correlated with the frequency of changes at a number of genetic loci (P<0.001), whereas CDK4 immunostaining showed negative correlation with 12q13.13 amplification. The genotypic similarity, at the limit of the array's resolution, between components implies that most genetic changes precede phenotypic ‘progression,’ early in tumorigenesis. The relationship between genetic changes and presentation or grade may reflect differences in factors that control genomic instability or the background genotype of the tumor.
Similar content being viewed by others
Main
Liposarcoma is one of the most common soft-tissue sarcomas in adults. The most common subtype consists of the well-differentiated–dedifferentiated family of liposarcomas. Well-differentiated liposarcoma is a tumor with a significant recurrence risk and mortality, but the most important risk factor for developing metastasis is the presence of ‘dedifferentiation,’ a relatively rare event.1, 2 Histologically, dedifferentiated liposarcoma consists of a biphasic tumor with both lipogenic (‘well-differentiated’) and non-lipogenic (‘dedifferentiated’) components.1, 2 In most instances, the latter component consists of a high-grade sarcoma, such as the so-called ‘malignant fibrous histiocytoma.’ However, in rare cases, the dedifferentiated area takes the form of a low-grade tumor resembling, for example, desmoid-type fibromatosis.3 Clinically, the dedifferentiated component can present either synchronously with the well-differentiated liposarcoma or metachronously many years after the initial diagnosis of a well-differentiated liposarcoma.3, 4, 5 Consequently, the dedifferentiated phenotype may represent the progression of a well-differentiated liposarcoma through stepwise accumulation of genetic changes.
The gene copy-number changes in most sarcomas preferentially involve additions rather than deletions, implying amplification of proto-oncogenes rather than loss of tumor-suppressor genes. Cytogenetic studies of liposarcomas have shown characteristic chromosomal rearrangements. More specifically, these tumors frequently show giant marker chromosomes and supernumerary ring chromosomes containing sequences derived from 12q, and less frequently from 1q.6, 7, 8, 9 Amplifications of genes MDM2, HMGA2, SAS and CDK4, located in the chromosomal subregion 12q13–q15 have been implicated in liposarcoma tumorigenesis.10, 11, 12, 13, 14 The changes in 12q, especially the amplification of MDM2 and CDK4, have been diagnostically useful in discriminating well-differentiated liposarcomas from benign lipomas and dedifferentiated liposarcomas from other high-grade sarcomas, respectively.13, 15, 16 In fact, improved understanding of liposarcoma genetics has supported the reclassification of some tumors from undifferentiated categories to dedifferentiated liposarcoma.17, 18
However, the specific genetic changes that distinguish between well-differentiated liposarcoma and dedifferentiated liposarcoma are poorly understood. As a group, dedifferentiated liposarcomas show more complex chromosomal aberrations than do well-differentiated liposarcomas.19 Intriguingly, cDNA microarray analysis shows that the well-differentiated component of dedifferentiated liposarcoma clusters distinctly from purely well-differentiated liposarcoma. The above finding suggests that only a subset of well-differentiated liposarcomas have the capacity for progression.20 Comparative genomic hybridization (CGH) confirms the presence of 12q13–q15 amplification in dedifferentiated liposarcoma, but in an overall more complex genetic profile, than in well-differentiated liposarcoma. Additional common amplifications include 1q23, 12q24 and either 6q23 or 1p32.13 Analysis of the 6q23 amplicon shows MAP3K5 as a candidate gene that, when amplified, may inhibit lipogenic differentiation through JUN- and PPAR-γ-dependent pathways.21 Amplification of 1p32 may increase the amount of JUN itself. Recently, it has also been shown that overexpression of some AP-1 transcription factors may block adipogenesis in vitro and in vivo.22, 23 Taken together, these results support a role of MAP kinase and AP-1 signaling pathways in liposarcoma progression.
Nevertheless, the above model does not completely explain tumor progression for several reasons. First, in the aforementioned studies, up to a third of dedifferentiated liposarcomas did not show any of these amplifications (other than 12q13–q15). Furthermore, the 1q23, 6q23 and 1p32 amplicons are similar in almost all cases in which both components were analyzed separately, suggesting that these genetic changes may already be present in the well-differentiated component. Finally, the majority of cases in the above-mentioned studies consist of only the dedifferentiated area or a mixture of both components of a given tumor. Thus, the relative contributions of the changes at 1q23, 6q23 and 1p32 or other loci in a liposarcoma progression model remain to be determined. A systematic comparison of paired well-differentiated and dedifferentiated areas in a large number of tumors, including synchronous and metachronous types, may allow examination of the genomic divergence between components. Genetic changes unique to one component may, in turn, uncover mechanisms that contribute to progression.
In this study, we compare the genetic changes of paired well-differentiated and dedifferentiated components of 29 well-characterized dedifferentiated liposarcomas using the superior resolution of array CGH. We explore the association between genetic changes and clinical presentation (synchronous or metachronous) or histological grade of the dedifferentiated area. We correlate CDK4 and MDM2 overexpression by immunohistochemistry with the genomic alterations in the tumor.
Materials and methods
Case Selection
We initially identified 30 cases of dedifferentiated liposarcomas from the pathology archives at our institution. Of these, 29 resulted in informative array CGH analysis and were further studied. We recently reported more detailed clinicopathologic parameters of 25 of these cases.24 The diagnosis was based on light microscopic, radiographic and clinical features. The ‘gold standard’ of diagnosis in every case was the presence of a well-differentiated liposarcoma either adjacent to macroscopic areas of sarcoma without lipogenic differentiation (synchronous) or previously removed from the same anatomical site (metachronous). The grade of the dedifferentiated component was recorded on the basis of previously published criteria.5, 25
Microdissection and DNA Extraction
Microdissection and DNA extraction were performed as described previously.26, 27 Briefly, tumor DNA was extracted from unstained sections obtained from a formalin-fixed paraffin-embedded tissue. Desired areas (well differentiated and dedifferentiated) of the tumors were identified on the basis of hematoxylin and eosin-stained slides. The selected regions were manually dissected into the extraction buffer (10 mM Tris, 1.5 mM MgCl2, 50 mM KCl and 0.5% Tween-20 (Fisher Scientific, Tustin, CA, USA) with 0.4 mg/ml proteinase K (Sigma, St Louis, MO, USA) and concentrated using Microcon YM-30 columns (Amicon Millipore, Bedford, MA, USA). DNA was quantitated as described previously.
DNA Amplification and Labeling
DNA was amplified and labeled according to published methods.28 Briefly, template DNA was random primed using the BioPrime DNA labeling system (Invitrogen, Carslbad, CA, USA) according to the manufacturer's directions. Amplified genomic DNA was labeled with cy3-dCTP, and reference genomic male DNA was labeled in the same manner with cy5-dCTP (Amersham Pharmacia, Piscataway, NJ, USA). Unincorporated fluorescent nucleotides were removed using Sephadex G-50 spin columns (Amersham Pharmacia).
Hybridization
Human scanning arrays consisting of 2464 bacterial artificial chromosome (BAC) clones spanning the genome at ∼1-Mb resolution were used.29, 30 Hybridization and washing were carried out as described previously.28 Labeled tumor DNA and reference DNA were mixed with Cot-1 DNA (Invitrogen) and hybridized to the array for 48 h at 37 °C in a humidified chamber with slow agitation. After washing, DNA was counterstained with 4′6-Diamidino-2-phenylindole (DAPI) in phosphate-buffered glycerol.
Image Analysis
The images, including fluorescence in the DAPI, Cy3 and Cy5 ranges were captured using a digitally charged-coupled device-based system. Image data were analyzed using the UCSF Cancer Center's Array Core SPOT and SPROC software.31
Statistical Considerations
All statistical analyses were carried out using the freely available R/Bioconductor software.32 The array CGH data were corrected for a geometrical dependence of the ratios on the array in some hybridizations, which translates into apparent measurement noise when ratios are plotted according to their positions in the genome using procedures described previously.33 The data were analyzed using circular binary segmentation to translate noisy intensity measurements into regions of equal copy numbers.34 Each probe was assigned a segment value referred to as its smoothed value. The scaled median absolute deviation of the difference between the observed and smoothed values was used to estimate the tumor-specific experimental variation. The gain and loss status for each probe was defined using the merged level procedure.35
The amplification status for a clone was determined by considering the width of the segment to which that clone belonged as well as a minimum difference between the smoothed value of the clone and the segment means of the neighboring segments. The clone was declared amplified if it belonged to the segment spanning <22 Mb, and the minimum difference was greater than , where x is the final smoothed value for the clone.
A clone-wise comparison of variables was made using moderated t-statistics.36 The false discovery rate (FDR) was controlled across all autosomes to obtain adjusted P-values, and a cutoff value of 0.05 was used to declare a clone to be significant. Paired t-tests were conducted to compare the matched dedifferentiated, well-differentiated components. To test the association of clinical presentation or grade, the effect was controlled as a covariate in a linear model. For analyzing the CDK4 and MDM2 interaction in the 12q13–15 region, a multiplicative model was fit for each clone and P-values were FDR adjusted for all clones in that region.
The paired Wilcoxon rank sum test was used to analyze the association of dedifferentiated, well-differentiated groups with summary genomic events, such as number of break points, number of chromosomes with break points, number of amplifications, number of chromosomes with amplifications, number of whole chromosome changes and fraction of genome altered. To test the association of summary genomic events with other phenotypes in the dedifferentiated, well-differentiated combined data set, a linear model was used in which the patient effect was controlled. The Wilcoxon rank sum test was used when testing within either the dedifferentiated or well-differentiated subset. A P-value cutoff of 0.05 was used to declare significance.
Unsupervised hierarchical clustering of the tumors was performed using smoothed imputed data with outliers present. Agglomerative hierarchical clustering with Pearson's correlation as a similarity measure and the Ward method to minimize the sum of variances was used to produce compact spherical clusters.
Immunohistochemistry
Immunohistochemical analysis was carried out using previously published techniques37 on archival, paraffin-embedded sections. Briefly, 4-μm paraffin-embedded sections were de-paraffinized, heated in EDTA buffer (Cell Marque, Rocklin, CA, USA), blocked and incubated with a monoclonal antibody to CDK4 (DCS-31, Biosource, Camarillo, CA, USA) at 1:100 dilution or MDM2 (IF2, Zymed, South San Francisco, CA, USA) at 1:100 dilution. Detection for all antibodies used the Envision system (Dako, Carpinteria, CA, USA) according to the manufacturer's directions. Slides were scored as positive if >10% of tumor cells showed specific, strong, nuclear staining. Positive cases were stratified as follows:38
level 1: >10%, but <25% positive cells;
level 2: > 25%, but <50% positive cells;
level 3: > 50%, but <90% positive cells;
level 4: > 90%, positive cells.
Results
Clinicopathologic Characteristics
The clinicopathologic information is summarized in Table 1. The study consisted of 29 matched pairs of well-differentiated and dedifferentiated liposarcomas. The clinical and pathological findings of 25 tumors were reported previously.24 The most common presentation was a retroperitoneal mass (n=21, 72%) with the remaining cases distributed between the extremity, trunk, abdomen and spermatic cord. In 25 cases (86%), the dedifferentiated liposarcoma was present at the time of first diagnosis (synchronous), whereas in the remaining cases (n=4, 14%), a diagnosis of well-differentiated liposarcoma preceded the presentation of the dedifferentiated liposarcoma by 1–5 years (metachronous). Grossly, tumors averaged 18 cm in greatest dimension (range: 5–45 cm). Histologically, all cases showed a component of the well-differentiated liposarcoma ranging from ∼10 to 75% of the tumor present on slides. All of the dedifferentiated components were grossly apparent (>2 cm). The most common subtype of the well-differentiated component was lipoma-like (n=19, 66%) although sclerosing and inflammatory types were also identified. Eight cases (28%) were classified as low-grade dedifferentiation on the basis of cellularity and cytomorphologic features more compatible with fibromatosis than with high-grade sarcoma. The remaining cases (n=21, 72%) consisted of high-grade sarcoma. One high-grade component demonstrated meningothelial-like whorls, and one had osteosarcomatous differentiation. The remaining high-grade components were morphologically and immunophenotypically ‘undifferentiated’ with spindle cell (n=10, 35%), round cell (n=2, 7%) or pleomorphic (n=7, 24%) cytomorphology. A dense acute and chronic inflammatory infiltrate imparting a so-called ‘inflammatory MFH’ appearance was present in two pleomorphic cases. Regardless of grade, all of the dedifferentiated components showed, at least focally, > 5 mitoses per 10 high power (40 × ) fields.
Genomic Alterations in Liposarcomas
Representative array CGH profiles from tumor pairs are illustrated in Figure 1. Using unsupervised hierarchical clustering of genetic gains and losses, (Figure 2) all but two tumors clustered as well-differentiated–dedifferentiated pairs. More specifically, the well-differentiated component of case 26 clustered with the tumor pair from patient 25 (most closely associated with the dedifferentiated component of this pair), sharing high-level amplification at 2p, gains at 1q, 7q, and 16p and losses at 8p and 12p.
Array CGH profiles from representative pairs of dedifferentiated liposarcoma components. Copy-number changes relative to normal DNA are shown for each clone. Clones are ordered from chromosomes 1 to 22 and within each chromosome based on their map position. The X and Y chromosomes are not informative in these studies as the control DNA is not gender matched. (a) Whole genome profile of case 22 shows essentially identical genotype between components. (b) Whole genome profile of case 14 shows increased copy number of 1q21–23 only in the dedifferentiated component and 14q32 only in the well-differentiated component. (c) Whole genome profile of case 24 which shows the increased copy number of 1p31–32 and 1q21—23, as well as numerous other gains and losses specific to the dedifferentiated component. (d) Chromosome 12 profile of case 22 shows similarity at 12q13—15, which consists of two amplicons.
Unsupervised hierarchical cluster analysis of genome copy number of 29 dedifferentiated liposarcoma pairs. The liposarcomas are distributed along the horizontal axis and genome is along the vertical axis. Chromosomes 1–22 are represented by the black and gray bars, respectively, at left and patient numbers are along the top. Three major clusters based on genetic complexity (complex, simple and intermediate) are arbitrarily ordered from left to right. Increased copy number is indicated in green, decreased copy number in red and high-level amplifications in yellow. The upper color bars indicate clinicopathologic properties of the tumor. Color codes are at the right of the figure: for differentiation status, green indicates dedifferentiated and blue indicates well-differentiated; for grade green indicates high and blue indicates low; for presentation green indicates synchronous and blue indicates metachronous. WD=well-differentiated component; DD=dedifferentiated component; 1o=primary; 2o=secondary.
The pattern of amplification at 12q13–15 was remarkably similar for a given tumor pair (Figure 1d). All 29 (100%) dedifferentiated liposarcomas had high-level amplifications in 12q13–15 in at least one component (Figure 1, Table 1). The 12q15 amplicon (BACs CTB-136O14 and CTB-82N15, including MDM2 at 64.5 Mb) was slightly more common (25/29, 86%) than was the 12q13–14 amplicon (BACs RMC12P001 (CDK4) and GS-56N1 (SAS) at 56.4 Mb) (23/29, 70%). Moreover, four tumors were discordant with respect to differentiation and amplification of these regions (Table 1). Contiguous amplification of the intervening region was uncommon (n=7, 24%), although gains in this region were sometimes present.
Differentiation-Specific Genomic Profiles
On the basis of cluster analysis and evaluation of tumor pairs, dedifferentiated areas showed more amplifications than did the matched well-differentiated components (P=0.008). However, no difference in whole chromosome changes or total genomic variability was observed. The only genetic change that showed a trend to specificity for the dedifferentiated component was the gain of 19q13.2, but statistical significance (P=0.003) was lost after adjusting for multiple comparisons. Thus, a hypothetical ‘differentiation-specific’ genetic change was not identified (Figure 3a). The 19q13.2 region corresponds to BACs RP11-21J15 and CTD-2271H24 (49.7–50.7 Mb), the location of the FOS-B gene.
Recurrent abnormalities in both components of 29 dedifferentiated liposarcoma pairs. Plots comparing the frequency of copy number increases (above the X axis) and decreases (below the X axis) of specific chromosomal regions on the basis of clinicopathologic parameters. Vertical lines indicate chromosome boundaries, and vertical dashed lines indicate centromeres. In each panel, the top two plots represent the fraction of cases with gains or losses as a function of genome location. The third plot shows the clone statistics (see ‘Materials and methods’ section). The green, blue and red horizontal dotted lines represent the threshold for P=0.1, 0.05 and 0.01, respectively (moderated t-test). (a) Well-differentiated compared with dedifferentiated components. (b) Low-grade dedifferentiated compared with high-grade dedifferentiated components. (c) Synchronous compared with metachronous clinical presentation.
Given previous reports associating amplifications of chromosomal regions 1p, 1q, 6q and 12q24 with dedifferentiated liposarcoma, we analyzed these regions in more detail (Table 1). Additional amplifications that were commonly observed in at least one component of the dedifferentiated liposarcomas included 1p31–32 (n=7, 24%), 1q21–23 (n=9, 31%), 6q23–24 (n=7, 24%) and 12q24 (n=3, 10%) (Table 1, Figures 1 and 2). Coamplification of 1p31–32 and 6q23–24 was a rare event but was detected in one case (Case 16, Table 1). The 1q21–23 and 6q23–24 amplicons were slightly more common in dedifferentiated components. (Table 1 and Figure 2). Again, these genetic changes did not achieve statistical significance with proper correction for multiple comparisons (Figure 3a), and the 6q23–24 and 1p31–32 amplicons were each identified in the well-differentiated, but not in the paired dedifferentiated, component of one case (Table 1).
In contrast, specific chromosomal changes associated with grade or clinical presentation. For example, the high-grade dedifferentiated components had significantly (P<0.001) more genomic gains at 6q14.1 (RP11-217L13, IRAK1BP1), 6q23.2–24.1(RP11-139O22 to RP11-15H7, MYB, MAP3K5), 10q21.1 (RP11-240K9 to RP11-133C15, PRGK) and 12q14.1–14.2 and losses at 8p21.3 (RP11-51C1, RP11-117P11 LPL) (Figure 3b). Similarly, comparison of synchronous with metachronous dedifferentiated liposarcomas showed that synchronous tumors had significantly more (P<0.001) gains at 6q23.2–24.1 (MYB, MAP3K5), 10q11.2–21.1 (PRGK) and 12q14.1–14.2, whereas metachronous tumors more often had losses at 8p21.3 (LPL) (Figure 3c).
CDK4 and MDM2 Status
The results of CDK4 and MDM2 immunohistochemistry are summarized in Table 1, and representative cases are shown in Figure 4. CDK4 was detected in 24 (83%) and 26 (89%) of the well-differentiated and dedifferentiated components, respectively, although the staining score was usually higher in the latter component within a given tumor (Table 1 and Figure 4). MDM2 was detected in 27 (93%) and 29 (100%) of the well-differentiated and dedifferentiated components, respectively. Again, the dedifferentiated component generally showed a higher staining score than did the paired well-differentiated component from the same case (Table 1 and Figure 4). No significant correlation was noted in total genomic variability or number of amplifications and CDK4 or MDM2 status (data not shown). Immunohistochemistry for CDK4 and MDM2 correlated with the amplification of 12q13.3–14 (P<0.001) and 12q15 (P=0.015), respectively, although the association was not exclusive (Table 1). More specifically, two cases were positive for CDK4 and MDM2 by immunohistochemistry without high-level amplification of 12q13.3–14 and 12q15, respectively. The above cases had gains of the respective regions of chromosome 12 (Figure 2).
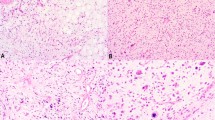
Immunohistochemical staining for CDK4 and MDM2 in dedifferentiated liposarcomas. Paired well-differentiated and dedifferentiated tumors are in the left and right columns, respectively. (a and b) Hematoxylin and eosin (H&E) of case 8. (c and d) MDM2 case 8. (e and f) H&E case 15. (g and h) CDK4 case 15.
Finally, we analyzed the interaction effect of the CDK4 and MDM2 immunophenotypes on the genome. In the well-differentiated components, this analysis showed a significant negative correlation effect of CDK4 immunostaining with 12q13.13 amplification. That is, MDM2+ CDK4− tumors had over sixfold more amplification of 12q13.13 (RP11-132H4) than did MDM2+ and CDK4+ tumors (P=0.002). This BAC maps ∼2.7 and 15 Mbp centromeric to the CDK4 and MDM2 gene loci, respectively, near the gene for transcription factor ATF7.
Discussion
The progression of liposarcoma from a well-differentiated to a dedifferentiated phenotype raises the possibility of specific genetic changes that mediate such progression. In this study, we tested the hypothesis that specific, reproducible genetic changes are responsible for the dedifferentiated phenotype. Using unsupervised hierarchical cluster analysis, paired well-differentiated and dedifferentiated liposarcomas segregated together in almost all cases. The results show that although dedifferentiated liposarcomas are a genetically complex group of tumors with many reproducible changes, no genetic change uniformly distinguishes the well-differentiated vs dedifferentiated components in a statistically significant manner. A trend for gain of 19q13.2 was observed in dedifferentiated components, but was only statistically significant if uncorrected for multiple comparisons. Nevertheless, the latter finding is intriguing because this region includes the FOS-B gene a member of the AP-1 family of transcription factors, which heterodimerizes with c-JUN to regulate transcription and may inhibit adipogenesis.23, 39 Thus, the finding of gains of the FOS-B gene is at least consistent with the importance of upregulated AP-1 signaling in the dedifferentiated phenotype.
These data support a strong clonal relationship between components of dedifferentiated liposarcoma, including metachronous tumors separated by as much as 5 years. In previous reports of expression array analyses, the well-differentiated components of dedifferentiated liposarcomas clustered distinctly from purely well-differentiated liposarcomas.20 Other studies have suggested that purely well-differentiated liposarcomas are genetically simpler and have distinct profiles from the well-differentiated components of dedifferentiated liposarcomas.13 Taken together, these findings imply that genetic changes mediating dedifferentiation may precede phenotypic changes. This conclusion is important because it indicates that the background genotypes of some apparently ‘well-differentiated’ liposarcomas already harbor the changes required for progression. In turn, the genetic changes that separate purely well-differentiated from dedifferentiated liposarcomas may be prognostically useful in predicting progression. Metachronous tumors are rare, and prospective studies may be necessary to confirm this hypothesis. Finally, we acknowledge that although the BAC-based array used in this study offers superior resolution to chromosomal CGH, at ∼1-Mb resolution, the BAC array is limited compared with some oligonucleotide-based platforms.40 The BAC array was chosen in this study because of consistent results when studying DNA from archival formalin-fixed paraffin-embedded tissue. However, these data do not rule out that small chromosomal changes, below the level of detection of the BAC array, or allelic imbalances, may distinguish well-differentiated and dedifferentiated components of liposarcoma.
A relationship between the amplification of specific loci (1p31–32, 1q21–23, 6q23–24 and 12q24) and the differentiation status of liposarcoma has been implicated in previous reports.13, 21, 41, 42 Some of these studies have also suggested that the 1p32 and 6q23 amplicons are mutually exclusive. That is, dedifferentiated liposarcomas demonstrate one, but not both, amplicons resulting in the overexpression of either JUN or MAP3K5 (which map to 1p32 and 6q23, respectively).13 Intriguingly, in a separate study, gene expression profiling comparing well-differentiated and dedifferentiated liposarcomas did not show an upregulation of either JUN or MAP3K5 in the top 50 differentially expressed genes.20 In the present study, amplification at each of these regions was relatively common, but was present in both the well-differentiated and dedifferentiated components. Interestingly, the 1p31–32 and 6q23–24 amplicons were occasionally identified only in the well-differentiated components. Only one tumor showed coamplification of 1p32 and 6q23 supporting the fact that, in the majority of tumors, amplification of these regions is mutually exclusive. Taken together, these results suggest that the above-mentioned amplicons are neither necessary nor sufficient to account for the phenotypic difference between well-differentiated and dedifferentiated components. However, the present findings are still consistent with a role of genes in the AP-1 signaling pathway (specifically, MAP3K5 and JUN) in dedifferentiated liposarcoma tumorigenesis. These data suggest that gene amplification may be one of multiple mechanisms (possibly also including point mutation, transcriptional regulation or protein stability) that modulate the AP-1 signaling pathway in liposarcoma.
Our data confirm that 12q13–q15 amplification is a very common event in both components of dedifferentiated liposarcoma.6, 7, 8, 9 The copy-number profile of the 12q13–q15 amplicon did not differ significantly between paired components, implying that quantitative or qualitative changes in this genomic segment do not contribute to the dedifferentiated phenotype. The array CGH results confirm recent findings that the amplification of 12q13–15 actually consists of two amplicons.14 More specifically, the 12q13.3–12q14 amplicon, which includes CDK4 and SAS, is discontinuous with the 12q15 amplicon (the location of MDM2). Contiguous amplification of the intervening region was a rare event in this study. Our results also support previous findings that MDM2 is more commonly amplified than is CDK4 in dedifferentiated liposarcomas.14, 41, 43 Nevertheless, MDM2 amplification is not universal as it was absent in at least one component of four cases. The very close correlation between CDK4 or MDM2 immunostaining and the 12q13.3–12q14 or 12q15 amplicons, respectively, is in keeping with the established diagnostic utility of these immunostains.15 A small number of cases showed positive immunostaining for either MDM2 or CDK4 without amplification of the corresponding gene, suggesting that other mechanisms that alter expression may be involved.44 The statistical analysis of the interaction effect of MDM2 or CDK4 immunostaining on 12q alterations showed an unexpected result, namely that MDM2+, CDK4− tumors had significantly high-level amplifications of a region centromeric to both MDM2 and CDK4. The BAC in question maps near the ATF7 gene, suggesting that this transcription factor and CDK4 may have redundant functions in promoting liposarcoma tumorigenesis.
Amplification of 1q21–23 has been identified in both well-differentiated and dedifferentiated liposarcomas.13 This is a relatively large segment; therefore, the exact genes involved are not known although COAS1-3 genes have been implicated in the past.45 In this study, the pattern of amplification within 1q21–23 varied between tumors. However, the most commonly amplified BAC within this region (CTD-2096I16 at 161.3–161.5 Mb) encompasses the RGS4 and RGS5 genes, ∼19 Mb telomeric to the COAS3 gene. RGS4 and RGS5 genes encode regulators of G-protein-coupled receptors. These RGS proteins negatively regulate G-protein-coupled receptor signaling which, in turn, mediates numerous metabolic and developmental processes.46 Obviously, the exact role and importance of these genes in liposarcoma will require additional studies.
This study identified genetic gains at a number of loci including PRGK, MYB, MAP3K5 and loss of LPL (the gene encoding lipoprotein lipase) that were significantly more common in high-grade and metachronous tumors as compared with low-grade and synchronous tumors, respectively. To our knowledge, these data are the first to specifically describe that the grade and presentation of the dedifferentiated component correlate with specific genetic changes. Little is known about the clinical significance of the grade of dedifferentiation, although some studies suggest that outcome is independent of tumor grade.3, 47 However, this observation is based on a relatively small number of patients, and as most cases occur in the retroperitoneum, local tumor effects, rather than metastasis, often influence outcome. The identification of specific regions of the genome that correlate with grade may provide insights into mechanisms of liposarcoma biology. Similarly, the changes specific to metachronous tumors may stem from differences in tumor initiation, factors that control genomic instability or the background genotype of the well-differentiated component. Insofar as metachronous tumors represent documented clinical progression, over time, of a well-differentiated liposarcoma into a non-lipogenic sarcoma, prospective studies of phenotypically well-differentiated liposarcomas may be the ideal model system to further study genes that influence tumor progression.
References
Fletcher CDM, Unni KK, Mertens F, (eds). Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, 2002, pp, 173–174.
Weiss SW, Goldblum JR . Enzinger and Weiss's Soft Tissue Tumors. Mosby: St. Louis, 2008.
Henricks WH, Chu YC, Goldblum JR, et al. Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol 1997;21:271–281.
Meis JM . ‘Dedifferentiation’ in bone and soft-tissue tumors. A histological indicator of tumor progression. Pathol Annu 1991;26 (Pt 1):37–62.
Weiss SW, Rao VK . Well-differentiated liposarcoma (atypical lipoma) of deep soft tissue of the extremities, retroperitoneum, and miscellaneous sites. A follow-up study of 92 cases with analysis of the incidence of ‘dedifferentiation’. Am J Surg Pathol 1992;16:1051–1058.
Dal Cin P, Kools P, Sciot R, et al. Cytogenetic and fluorescencein situhybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet Cytogenet 1993;68:85–90.
Pedeutour F, Suijkerbuijk RF, Van Gaal J, et al. Chromosome 12 origin in rings and giant markers in well-differentiated liposarcoma. Cancer Genet Cytogenet 1993;66:133–134.
Mertens F, Dal Cin P, De Wever I, et al. Cytogenetic characterization of peripheral nerve sheath tumours: a report of the CHAMP study group. J Pathol 2000;190:31–38.
Suijkerbuijk RF, Olde Weghuis DE, Van den Berg M, et al. Comparative genomic hybridization as a tool to define two distinct chromosome 12-derived amplification units in well-differentiated liposarcomas. Genes Chromosomes Cancer 1994;9:292–295.
Dei Tos AP, Doglioni C, Piccinin S, et al. Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol 2000;190:531–536.
Pedeutour F, Suijkerbuijk RF, Forus A, et al. Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer 1994;10:85–94.
Pilotti S, Della Torre G, Lavarino C, et al. Molecular abnormalities in liposarcoma: role of MDM2 and CDK4-containing amplicons at 12q13-22. J Pathol 1998;185:188–190.
Chibon F, Mariani O, Derre J, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet 2002;139:24–29.
Italiano A, Bianchini L, Keslair F, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer 2008;122:2233–2241.
Binh MB, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol 2005;29:1340–1347.
Weaver J, Downs-Kelly E, Goldblum JR, et al. Fluorescencein situhybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol 2008;21:943–949.
Coindre JM, Hostein I, Maire G, et al. Inflammatory malignant fibrous histiocytomas and dedifferentiated liposarcomas: histological review, genomic profile, and MDM2 and CDK4 status favour a single entity. J Pathol 2004;203:822–830.
Coindre JM, Mariani O, Chibon F, et al. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol 2003;16:256–262.
Rosai J, Akerman M, Dal Cin P, et al. Combined morphologic and karyotypic study of 59 atypical lipomatous tumors. Evaluation of their relationship and differential diagnosis with other adipose tissue tumors (a report of the CHAMP Study Group). Am J Surg Pathol 1996;20:1182–1189.
Shimoji T, Kanda H, Kitagawa T, et al. Clinico-molecular study of dedifferentiation in well-differentiated liposarcoma. Biochem Biophys Res Commun 2004;314:1133–1140.
Chibon F, Mariani O, Derre J, et al. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes Chromosomes Cancer 2004;40:32–37.
Mariani O, Brennetot C, Coindre JM, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 2007;11:361–374.
Sabatakos G, Sims NA, Chen J, et al. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med 2000;6:985–990.
Horvai AE, Schaefer JT, Nakakura EK, et al. Immunostaining for peroxisome proliferator gamma distinguishes dedifferentiated liposarcoma from other retroperitoneal sarcomas. Mod Pathol 2008;21:517–524.
Folpe AL, Chand EM, Goldblum JR, et al. Expression of Fli-1, a nuclear transcription factor, distinguishes vascular neoplasms from potential mimics. Am J Surg Pathol 2001;25:1061–1066.
Gong G, DeVries S, Chew KL, et al. Genetic changes in paired atypical and usual ductal hyperplasia of the breast by comparative genomic hybridization. Clin Cancer Res 2001;7:2410–2414.
Waldman FM, DeVries S, Chew KL, et al. Chromosomal alterations in ductal carcinomasin situand theirin siturecurrences. J Natl Cancer Inst 2000;92:313–320.
Hwang ES, Nyante SJ, Yi Chen Y, et al. Clonality of lobular carcinomain situand synchronous invasive lobular carcinoma. Cancer 2004;100:2562–2572.
Hodgson G, Hager JH, Volik S, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet 2001;29:459–464.
Snijders AM, Fridlyand J, Mans DA, et al. Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene 2003;22:4370–4379.
Jain AN, Tokuyasu TA, Snijders AM, et al. Fully automatic quantification of microarray image data. Genome Res 2002;12:325–332.
R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2005.
Gajduskova P, Snijders AM, Kwek S . et al. Genome position and gene amplification. Genome Biol 2007;8:R120.
Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557–572.
Willenbrock H, Fridlyand J . A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics 2005;21:4084–4091.
Smyth GK . Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article 3.
Horvai AE, Kramer MJ, O’Donnell R . Beta-catenin nuclear expression correlates with cyclin D1 expression in primary and metastatic synovial sarcoma: a tissue microarray study. Arch Pathol Lab Med 2006;130:792–798.
Horvai AE, Li L, Xu Z, et al. c-Kit is not expressed in malignant mesothelioma. Mod Pathol 2003;16:818–822.
Castellazzi M, Spyrou G, La Vista N, et al. Overexpression of c-jun, junB, or junD affects cell growth differently. Proc Natl Acad Sci USA 1991;88:8890–8894.
Ylstra B, van den Ijssel P, Carvalho B, et al. BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH). Nucleic Acids Res 2006;34:445–450.
Hostein I, Coindre JM, Derre J, et al. Comparative genomic hybridization study of paraffin-embedded dedifferentiated liposarcoma fixed with Holland Bouin's fluid. Diagn Mol Pathol 2003;12:166–173.
Rieker RJ, Joos S, Bartsch C, et al. Distinct chromosomal imbalances in pleomorphic and in high-grade dedifferentiated liposarcomas. Int J Cancer 2002;99:68–73.
Heidenblad M, Hallor KH, Staaf J, et al. Genomic profiling of bone and soft tissue tumors with supernumerary ring chromosomes using tiling resolution bacterial artificial chromosome microarrays. Oncogene 2006;25:7106–7116.
Deb SP . Function and dysfunction of the human oncoprotein MDM2. Front Biosci 2002;7:d235–d243.
Nilsson M, Meza-Zepeda LA, Mertens F, et al. Amplification of chromosome 1 sequences in lipomatous tumors and other sarcomas. Int J Cancer 2004;109:363–369.
Hendriks-Balk MC, Peters SLM, Michel MC, et al. Regulation of G protein-coupled receptor signalling: focus on the cardiovascular system and regulator of G protein signalling proteins. Eur J Pharmacol 2008;585:278–291.
Elgar F, Goldblum JR . Well-differentiated liposarcoma of the retroperitoneum: a clinicopathologic analysis of 20 cases, with particular attention to the extent of low-grade dedifferentiation. Mod Pathol 1997;10:113–120.
Acknowledgements
We would like to thank the Immunopathology Laboratory at UCSF for their technical assistance. This research was supported by the UCSF Research Evaluation and Allocation Committee (REAC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Horvai, A., DeVries, S., Roy, R. et al. Similarity in genetic alterations between paired well-differentiated and dedifferentiated components of dedifferentiated liposarcoma. Mod Pathol 22, 1477–1488 (2009). https://doi.org/10.1038/modpathol.2009.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.119
Keywords
This article is cited by
-
Co-expression of MDM2 and CDK4 in transformed human mesenchymal stem cells causes high-grade sarcoma with a dedifferentiated liposarcoma-like morphology
Laboratory Investigation (2019)
-
A challenging diagnosis of mesenchymal neoplasm of the colon: colonic dedifferentiated liposarcoma with lymph node metastases—a case report and review of the literature
International Journal of Colorectal Disease (2019)
-
Integrated exome and RNA sequencing of dedifferentiated liposarcoma
Nature Communications (2019)
-
Scattered genomic amplification in dedifferentiated liposarcoma
Molecular Cytogenetics (2017)
-
Genomic profiling of malignant phyllodes tumors reveals aberrations in FGFR1 and PI-3 kinase/RAS signaling pathways and provides insights into intratumoral heterogeneity
Modern Pathology (2016)