Abstract
High-mobility group A2 is highly expressed during embryogenesis and in various benign and malignant tumors. Recent studies report that high-mobility group A2 is negatively regulated by the let-7 microRNAs (miRNAs) family in vitro. The development of pituitary adenomas in high-mobility group A2 transgenic mice showed that high-mobility group A2 may be involved in pituitary tumorigenesis. However, no studies have investigated the clinical significance of high-mobility group A2 and its relationship to the let-7 miRNA family in human pituitary adenomas. Using immunohistochemistry, we analyzed high-mobility group A2 expression with respect to various clinicopathologic factors in 98 pituitary adenomas. Overexpression of high-mobility group A2 was observed in 39% (38/98) of pituitary adenomas compared with normal adenohypophysial tissue and was frequently found in adenomas including prolactin (PRL), adrenocorticotrophic hormone, or follicle-stimulating hormone/luteinizing hormone and in null cell adenomas, but relatively rare in growth hormone (GH) and mixed GH/PRL adenomas. High-mobility group A2 expression was significantly associated with tumor invasion (P<0.05) and was significantly higher in grade IV than in grades I, II, and III adenomas (P<0.05). High levels of high-mobility group A2 expression were more frequently observed in macroadenomas than in microadenomas (P<0.05). High levels of high-mobility group A2 expression also significantly correlated with the proliferation marker Ki-67 (P<0.0001). Real-time quantitative RT-PCR analysis was carried out to evaluate the expression of let-7 in 55 pituitary adenomas. Subsequently, decreased expression of let-7 was confirmed in 23 of 55 (42%) adenomas and was correlated with high-grade tumors (P<0.05). An inverse correlation between let-7 and high-mobility group A2 expression was evident (R=−0.33, P<0.05). These findings support a causal link between let-7 and high-mobility group A2 whereby loss of let-7 expression induces high-mobility group A2 upregulation that represents an important mechanism in pituitary tumorigenesis and progression.
Similar content being viewed by others
Main
Pituitary adenomas are common intracranial neoplasms, comprising 10–15% of diagnosed brain tumors.1 They may arise from any of the five differentiated cell types within this gland: somatotropes, lactotropes, corticotropes, thyrotropes, and gonadotropes, which secrete growth hormone (GH), prolactin (PRL), adrenocorticotrophic hormone (ACTH), thyroid-stimulating hormone (TSH), and gonadotropins (follicle-stimulating hormone (FSH) and luteinizing hormone (LH)), respectively. Pituitary adenomas are usually biologically benign, but often associated with specific endocrine syndromes, such as acromegaly, amenorrea–galactorrea, Cushing disease, TSH-induced hyperthyroidism, and hypopituitarism. The tumors can also cause tumor mass effect (ie, local compressive effects on brain structures and cranial nerves). The identification of molecular pathways leading to pituitary tumorigenesis represents one of the major challenges in endocrine oncology. Several factors, such as gene mutations, hypothalamic dysregulation, and locally produced growth factors, appear to be, though controversial, involved in the transformation of pituitary cells.2
The high-mobility group A (HMGA) proteins are nonhistone chromosomal proteins that bind through their AT-binding motifs to the minor groove of AT-rich DNA strands. They have no intrinsic transcriptional activity but can modulate transcription by altering chromatin architecture.3, 4, 5 High-mobility group A2 (HMGA2) proteins are widely expressed during embryogenesis, whereas their expression is low or absent in normal adult tissues.6, 7 HMGA2 proteins are involved in many diverse biological processes such as regulation of transcription, embryogenesis, differentiation, neoplastic transformation, and integration and expression of viral genomes.8 HMGA2 overexpression is a hallmark of various benign and malignant tumors and is also associated with a highly malignant phenotype and is a poor prognostic index.9 It was reported that transgenic mice overexpressing HMGA2 develop pituitary adenomas secreting PRL and GH and may be involved in pituitary tumorigenesis.10 However, HMGA2 expression was just studied in a small series of human sporadic prolactinomas and nonfunctional pituitary adenomas.11, 12 To date there has been no study of HMGA2 expression in a large series of human pituitary adenomas including all subtypes and its relationship to clinicopathological features in these tumors.
MicroRNAs (miRNAs) represent an emerging class of small endogenous noncoding RNAs (approximately 18–24 nucleotides) and they may regulate gene expression at the posttranscription level by direct cleavage of a target mRNA using interference machinery (mRNA cleavage) or by inhibition of protein synthesis.13, 14 miRNAs have been implicated in a variety of developmental and physiological processes, including cell proliferation and differentiation, apoptosis, metabolism, and morphogenesis.15 Recent evidence has shown that miRNA misexpression correlates with various human cancers and indicates that some miRNAs can function as oncogenes or tumor suppressors.14 Let-7 was first identified in Caenorhabditis elegans and is highly conserved in C. elegans, Drosophila, zebrafish, and humans.16, 17, 18 Let-7 is barely detectable in embryonic stages but increases in differentiated and mature tissues.18 Let-7 was indicated as a tumor suppressor in lung and colon cancer through targeting of RAS oncogene.19 Recently, several studies reported that let-7 can negatively regulate HMGA2 expression in a mouse model system,20 in head and neck cancers,21 in uterine leiomyomas,22 in lung cancer cell lines,23 and in ovarian cancers.24 However, very few data are available concerning the role of miRNA in pituitary physiology and disease and there have been no reports investigating a possible correlation between HMGA2 and the let-7 miRNA in pituitary adenomas.
In this study, we investigated the expression of HMGA2 in a series of 98 human pituitary adenomas of the various types and in 4 normal pituitary glands and compared these data with tumor type, size, invasiveness, and the labeling index of the proliferation marker Ki-67 antigen. We also investigated possible associations between HMGA2 expression and let-7 miRNA expression in pituitary adenomas.
Materials and methods
Human Normal and Pituitary Adenoma Samples
Normal human adenohypophyses were obtained at autopsy from four patients without endocrine dysfunction at Tokushima University Hospital (Tokushima, Japan). These tissues were examined using the hematoxylin–eosin stain and immunocytochemistry to exclude the possibility of incidental tumors. Ninety-eight postsurgical pituitary adenoma tissue samples were obtained from Tokushima University Hospital and Toranomon Hospital (Tokyo, Japan). These included 28 somatotroph, 5 mammosomatotroph, 16 lactotroph, 18 corticotroph (8 associated with Cushing's disease and 10 silent), 3 thyrotroph, 22 gonadotroph, 3 silent subtype-3, and 3 null cell adenomas (Table 1). Tumor size and invasiveness were defined on the basis of preoperative radiological investigations and operative findings, with a modified Hardy's classification.25 Grade I (microadenomas, <1 cm in diameter) and grade II (enclosed macroadenomas with or without suprasellar extension, ≥1 cm in diameter) tumors were defined as noninvasive. Grade III (local invasion of sphenoid and/or cavernous sinus) and grade IV tumors (with central nervous system/extracranial spread with or without metastasis) were considered to be invasive. Thus, 98 tumors included 11 tumors of grade I, 43 tumors of grade II, 32 tumors of grade III, and 12 tumors of grade IV (54 noninvasive and 44 invasive adenomas, Tables 1 and 3). None of the tumors examined in this study had evidence of postoperative recurrence.
Immunohistochemical
To detect HMGA2 protein and Ki-67 antigen, immunolocalization experiments were carried out on sections from representative blocks of paraffin-embedded tissues using the labeled streptavidin–biotin method, as previously described.26, 27 After deparaffinization and antigen retrieval using an autoclave oven technique, sections were incubated at 4°C overnight with goat polyclonal anti-HMGA2 antibody (1:50; HMGI-C, S-15, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or with Ki-67 antigen mouse monoclonal antibody (1:100; DakoCytomation, Glostrup, Denmark). Antigen–antibody complexes were detected using the cobalt-3,3′-diaminobenzidine reaction. The slides were counterstained lightly with hematoxylin or 1% methyl green and mounted for microscopic examination. A squamous cell carcinoma known to be positive for HMGA2 was used as a positive control. Sections incubated in PBS without the primary antibody served as negative controls.
Each slide was examined by an observer blinded to the diagnosis and clinicopathologic data; and reviewed and confirmed by a second blinded observer. Any intensity of nuclear staining was considered to present a positive stain for HMGA2 and Ki-67. A total of 500–1000 cells were counted and the percentage of HMGA2-stained tumor cells was scored on a scale of 0–4 (0, no staining; 1+, 1–20%; 2+, 20–50%; 3+, 50–80%; 4+, >80%). Furthermore the expression level of HMGA2 was divided as following three groups: negative, (0); moderate, (1+, 2+); high, (3+, 4+).
The Ki-67 antigen labeling index was determined by counting the number of positive cells in a total of 500–1000 tumor cells observed in several representative high-power fields ( × 400). The results were expressed as percentage of tumor cells with positive nuclei.
RNA Isolation and Real-Time Quantitative RT-PCR Analysis
Total RNA isolation and enrichment of small RNAs were performed using a mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer's protocol. For the miRNA quantitative RT-PCR (qRT-PCR) analysis, enriched small RNAs from 4 normal human adenohypophysis and 55 human pituitary adenomas were used. All samples were frozen and stored at −80°C. Because let-7 family has similar function, we used a primer set that may amplify multiple let-7 family members. The expression of human mature let-7 (hsa-let-7) miRNA was analyzed using a mirVana qRT-PCR miRNA detection kit (Ambion catalog nos. AM1558 and AM30000) according to manufacturer's protocol. Ubiquitously expressed U6 small nuclear RNA (Ambion catalog no. AM30303) was used for normalization. HeLa cell was chosen as positive control for let-7 expression. Briefly, reverse transcription reaction was performed with 20 ng of total RNA using gene-specific RT primers for let-7 and U6, respectively. Real-time PCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems), in a 25 μl reaction mixture consisting of 10 μl reverse transcription product, 5 μl mirVana 5 × Buffer, 0.5 μl 50 × ROX, 0.5 μl mirVana PCR primers and thermostable DNA polymerase (1 unit). PCR cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Each sample was analyzed in duplicate. Serial dilutions of cDNA from a pool of three normal pituitaries were used for the standard curve calculation. The standard curve method was used to determine relative quantitation of let-7. The relative amount of let-7 in each sample was normalized to the U6 RNA. The fold change values indicate the relative change in the expression levels between tumors samples and normal pituitary tissues assuming that the value of normal sample was equal to 1. The comparative Ct method (2−ΔΔCt) was also be used to confirm the results from the standard curve method.28, 29
Statistical Analyses
To determine the significance of associations between different variables, data were statistically analyzed by Mann–Whitney U-test, Kruskal–Wallis test, χ2-test, Spearman's correlation coefficient, and Pearson's correlation coefficient, using StatView J-4.5 software (Abacus Concepts, Berkeley, CA, USA). A P-value of less than 0.05 was considered statistically significant.
Results
Overexpression of HMGA2 Oncoprotein in Human Pituitary Adenomas
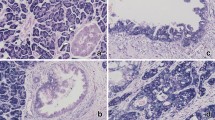
In this study, we examined HMGA2 expression in four normal pituitary glands and the 98 adenoma specimens. There was no appreciable immunostaining of HMGA2 in all normal pituitary tissues tested (Figure 1a). A clear nuclear staining for HMGA2, without membrane or cytoplasmic localization, was detected in 38 (39%) pituitary adenomas (Figure 1b–d; Table 2). HMGA2 immunoreactivity was frequently observed in three subtypes of pituitary adenomas; FSH/LH cell adenomas (15 of 22 cases, 68%), ACTH cell adenomas (12 of 18 cases, 67%), and PRL cell adenomas (5 of 16 cases, 31%). However, HMGA2 immunoreactivity was found rarely in GH cell adenomas (2 of 26 cases, 7%) and was not found in mixed GH/PRL adenomas (P<0.0001, Table 2).
Detection of HMGA2 immunoreactivity in pituitary normal tissues and 98 adenomas. (a) In normal pituitary cells HMGA2 showed negative immunoreaction. In contrast to normal cells, positive nuclear staining of HMGA2 was observed in tumor cells such as prolactinoma (3+; b), silent ACTH cell adenoma (4+; c) and FSH/LH cell adenoma (4+; d). The immunoreactivity of HMGA2 is variable but always very intense throughout the nuclei. The differences in immunostaining scores of HMGA2 between invasive and noninvasive adenomas or macroadenomas and microadenomas or among tumor grades were analyzed, respectively. (e) HMGA2 expression levels were significantly higher in invasive pituitary adenomas than in noninvasive pituitary adenomas (P<0.05, Mann–Whitney U-test). (f) HMGA2 expression levels were potentially higher in macroadenomas than in microadenomas (P=0.36, Mann–Whitney U-test). (g) HMGA2 expression levels were significantly correlated with tumor grade (P<0.05, Kruskal–Wallis test). (h) Ki-67 labeling index levels were significantly correlated with HMGA2 expression (P<0.05, Kruskal–Wallis test). Immunoreactivity score: 0, no staining; 1+, 1–20%; 2+, 20–50%; 3+, 50–80%; 4+, >80%. Negative, scale 0; moderate, scales 1+ and 2+; high, scales 3+ and 4+.
Expression of HMGA2 as high level and moderate level was detected in 24 of 98 and 14 of 98 adenomas, respectively. Of 98, 60 adenomas showed negative expression of HMGA2 (Table 3). The expression of HMGA2 was significantly higher (P<0.05, Figure 1e) and tended to be present with greater frequency in invasive adenomas than in noninvasive adenomas (Table 3). In addition, the expression of HMGA2 was more frequent (P<0.05, Table 3) and tended to be higher in macroadenomas than in microadenomas (Figure 1f). The overall level of HMGA2 expression was significantly higher in grade IV adenomas than grades I, II, and III (P<0.05, Figure 1g). Also high-level expression of HMGA2 case was frequently detected in grade IV (P<0.05, Table 3). Furthermore, Ki-67 labeling index correlated with HMGA2 overexpression (R=0.395, P<0.0001) and progressively increased from HMGA2 negative expression cases to moderate expression cases and high expression cases (P<0.0001, Figure 1h). The overexpression of HMGA2 was not related to patient age and gender (data not shown).
Aberrant Let-7 Expression in Human Pituitary Adenomas
Real-time qRT-PCR method was used to address the levels of let-7 in 4 normal and 55 adenomatous pituitary samples. As shown in Figure 2a and Table 4, compared with normal tissues, the reduced expression of let-7 was found in 23 (41.8%) adenomas (most cases exhibited >50% reduction). Slight upregulation of let-7 expression levels was also observed in the 15 (27%) adenomas (Figure 2a; Table 4; upregulation rates were less than 3-fold, most cases showed 1 to 2-fold level expression), whereas high upregulation of let-7 was detected in remaining 17 (31%) cases (upregulation rates were over 3-fold, few cases showed over 10-fold level expression). HeLa cells showed high level of let-7 (upregulation rate, fourfold). These aberrant expressions of let-7 may have tumor type–specificity. Reduction of let-7 was frequently detected in three subtypes of pituitary adenomas: PRL cell adenomas (6 of 9, 68%), ACTH cell adenomas (7 of 12, 58%), and FSH/LH cell adenomas (6 of 17 cases, 35%). However, in GH cell adenomas, reduction of let-7 was found only in 1 of 12 (8%) cases, whereas high upregulation was observed in 6 of 12 (50%) cases (Table 4).
Let-7 expression and HMGA2 immunostaining profile analyzed in 55 pituitary adenomas. (a) The value of let-7 expression in normal pituitary tissues was equal to onefold. The reduced expression of let-7 was found in 23 (41.8%) adenomas (blue square columns, downregulation rates were −1.10 to −16.41-fold, 17 cases showed over 2-fold change). Slight upregulation of let-7 was also observed in the 15 (27.2%) adenomas (red square columns, upregulation rates were less than 3-fold, 1.06 to 2.98-fold, 9 cases showed 1 to 2-fold level expression), whereas high upregulation of let-7 was detected in remaining 17 (31%) cases (green square columns, upregulation rates were over 3-fold, 3.24 to 22.21-fold, 5 cases showed over 10-fold level expression). In addition, 25 of 55 adenomas showed HMGA2 immunoactivity (purple square columns, in this figure, immunoreactivity score was converted to fold change temporarily: 0=0-fold; 1+=1-fold; 2+=2-fold; 3+=3-fold; 4+=4-fold). HMGA2 immunostaining were detected in 14 of 23 let-7 downregulated adenomas, 7 of 15 in let-7 slightly upregulated adenomas and in 4 of 23 let-7 highly upregulated adenomas, respectively. (b) The reduction of let-7 expression was potentially related to tumor grade. In grade IV, tumors showed the lowest level of let-7 (P<0.05, Kruskal–Wallis test). Grade I and grade II adenomas were placed together because grade I has a small number of cases (n=2).
When analyzed the clinicopathological characteristics of 23 pituitary adenomas with reduced let-7 expression, there were no notable differences on patient age and sex (data not shown). Interestingly, the expression level of let-7 was significantly lower in high-grade (III, IV) adenomas than in low-grade (I and II) adenomas (Figure 2b), and potentially lower in invasive adenomas than in noninvasive adenomas, although the differences were not statistically significant (data not shown).
Inverse Correlation between Let-7 Expression and HMGA2 Expression
At first, the expression of HMGA2 was negative in all four normal pituitary tissues that showed abundant let-7 by RT-PCR (data not shown). We further investigated the relationship between let-7 expression and HMGA2 expression in 55 pituitary adenomas through several viewpoints. Generally, 25 of 55 adenomas showed HMGA2 immunoactivity (Figure 2a; Table 4). Pituitary adenomas with positive HMGA2 immunostaining were more frequently detected in let-7 downregulated adenomas (14 of 23, 61%) than in let-7 slightly upregulated adenomas (7 of 15, 47%) and let-7 highly upregulated adenomas (4 of 17, 24%; P<0.05, Figure 2a; Table 4). Notably, high-level expression of HMGA2 was not detected in let-7 highly upregulated adenomas (Table 4). In addition, HMGA2 expression levels were higher in let-7 downregulated adenomas than in let-7 slightly upregulated adenomas and let-7 highly upregulated adenomas (P<0.05, Figure 3a). On the other hand, significantly lower expression of let-7 was detected in pituitary adenomas with high HMGA2 expression (P<0.05, Figure 3b). Finally, this inverse correlation between the expression of let-7 and HMGA2 in human pituitary adenomas was confirmed using Pearson's correlation coefficient analysis (r=−0.33, P<0.05, Figure 3c) and Spearman's correlation coefficient analysis (P=0.003).
Let-7 expression was negatively associated with HMGA2 immunoreactivity in pituitary adenomas. (a) HMGA2 expression levels were significantly higher in let-7 downregulated adenomas than in let-7 slightly upregulated adenomas and highly upregulated adenomas (P<0.05, Kruskal–Wallis test). (b) The significantly lower expression of let-7 was detected in pituitary adenomas with high HMGA2 expression (P<0.05, Kruskal–Wallis test). (c) A significant inverse correlation between the expression of let-7 and HMGA2 in pituitary adenomas was demonstrated using Pearson’s correlation coefficient analysis (r=−0.33, P<0.05).
Discussion
A high expression of HMGA2 has been detected in many kinds of benign tumor and cancer. It is also associated with a highly malignant phenotype and is a poor prognostic index.9 As HMGA2 transgenic mice develop mixed GH-PRL cell pituitary adenomas,10 and upregulation of HMGA2 protein has been found in human prolactinomas and nonfunctioning adenomas,11, 12 HMGA2 is a reasonable candidate oncogene for involvement in pituitary tumorigenesis. To our knowledge, our study is the first report concerning the clinical significance of HMGA2 overexpression in pituitary adenomas. In this study, upregulated expression of HMGA2 protein was found in a significant fraction (39%) of pituitary adenomas and associated with prognostic factors such as tumor grade, extent of invasion, tumor size, and cell proliferation marker. A strong correlation existed between HMGA2 overexpression and tumor cell invasion has been detected in breast cancer and gastric cancer.30, 31 In oral squamous cell carcinomas, strong staining of HMGA2 and loss of E-cadherin expression were detected at the invasive front of tumor.32 Our previous study also demonstrated that tumor-specific downregulation of E-cadherin and H-cadherin related to invasiveness of pituitary adenoma.27 HMGA2 may be involved in tumor cell invasion because of association with epithelial–mesenchymal transition that triggers tumor cell invasion. HMGA2 may regulate transcription factors, such as Snail, Slug, Twist, and inhibitor of differentiation 2, resulting in repressing E-cadherin expression and are contributed to a response to TGF-β that causes epithelial–mesenchymal transition.33 In addition several studies have demonstrated that blocking of HMGA2 protein synthesis has a negative effect on tumor cell proliferation.10, 34 Cell growth and oncogenic activity of HMGA proteins are based on their ability to downregulate or upregulate the expression of genes such as E2F1, cyclin A, and p53 that have a crucial function in the control of cell proliferation.9 These observations supported our findings that HMGA2 overexpression is related to tumor development and invasiveness in pituitary adenomas.
Characterization of HMGA2 overexpression with clinicopathological data in a large series of pituitary adenomas including all major types were not reported. Specially, studies on expression of HMGA2 in GH cell and ACTH cell adenomas have not been reported. Interestingly, we observed preferable expression of the HMGA2 protein in either ACTH or FSH/LH cell adenomas, as compared to GH or mixed GH-PRL cell adenomas. This is inconsistent with the observations in HMGA2 transgenic models developing mixed GH-PRL cell adenomas.10 The reasons for this discrepancy were currently unknown, but the underlying oncogenic mechanisms of pituitary adenoma may vary depending on species and cell types. This tumor type-specific alteration also has been reported for some tumor suppressor genes, CDKN2A, RB1, and RASSF1A; and some oncogenes, Gsp and FGFR4.28, 35, 36, 37, 38 These observations indicate that different types of pituitary adenomas may have distinct etiologic factors and pathogenetic mechanisms.
Let-7 is one of the founding members of the miRNA family, and several lines of evidence suggest that human let-7 acts as a putative tumor suppressor. In human, various let-7 genes map to chromosomal regions altered or deleted in human tumors.39 Expression of let-7 has been found to be downregulate in lung, colon, and ovarian cancers,24, 40, 41 and its expression levels to be associated with cancer progression. In our study, at first, sequencing analysis of miRNA library implied that let-7 levels were downregulated in pituitary adenomas (data not shown); then using qRT-PCR, we demonstrated that let-7 expression was significantly decreased in more than one-third of the pituitary adenomas, an observation consistent with the previous microarray data from a small series of pituitary adenomas.42 We further found that reduced let-7 expression correlated with high tumor grade. These findings thus provide a clinical evidence supporting that let-7 may function as a tumor suppressor miRNA, involving in the development and progression of human pituitary adenomas.
Recently, several groups independently reported that expression of HMGA2 is suppressed by let-7 in vitro,20, 23 and that disrupting the let-7 regulation of the HMGA2 enhances oncogenic transformation.20 Intriguingly, human HMGA2 gene has been known as a frequent target of chromosomal rearrangements that cause the loss of the C terminus of the protein and the 3′-UTR of the mRNA, the latter of which contains putative let-7-binding sites.9 In prolactinomas, upregulation of HMGA2 may be associated with amplification and/or rearrangement of the HMGA2 locus; but, in contrast to prolactinomas, rearrangement of the HMGA2 locus was rare in nonfunctional pituitary adenomas with HMGA2 overexpression.9 In our study, we confirmed that there was a significant inverse correlation between let-7 and HMGA2 expressions in human pituitary adenomas. In addition to loss of let-7-mediated repression because of truncation of HMGA2, let-7 reduction itself also may contribute to HMGA2 overexpression. Our findings first addressed the clinical significance of the let-7/HMGA2 connection in a large series of human pituitary tumors.
However, other aberrant expression of let-7 also has been detected. Upregulation of let-7 has been demonstrated in pituitary adenomas and some of them showed very high level of let-7. Except for functioning as tumor suppressor, does let-7 have oncogenic function in pituitary tumorigenesis through repressing other targets? Oncogenic function of let-7a-3 has been reported in a lung cancer model.43 Furthermore, we could not always demonstrate the concerted expression changes of let-7 and HMGA2 in pituitary adenomas. This could be because of technical/interpretive difficulties, but is more likely because of other possibilities. In some let-7 downregulated adenomas, HMGA2 expression has not been observed. This phenomenon implied that reduced let-7 may not be only one initial reason contributed to HMGA2 overexpression in those cases; other miRNAs such as mir-98 may also relate to HMGA2 regulation in pituitary adenomas.21 We noted that HMGA2 expression has been rarely detected in adenomas with high levels of let-7. Similar finding also has been discussed in small leiomyomas.44 But HMGA2 expression has been detected in about half of adenomas with normal or a little high expression of let-7. These findings implied that more let-7 may be needed in tumor than in normal tissue for effectively regulating target genes.
In conclusion, we found no immunoreactivity for HMGA2 in normal adult anterior pituitary glands. Nuclear expression of HMGA2 is common in the major types of pituitary adenomas with the exception of GH or mixed GH-PRL cell adenomas. High levels of expression of HMGA2 correlated with tumor grade, extent of invasion, tumor size, and higher Ki-67 proliferation index. These data support the premise that HMGA2 overexpression is an important and frequent event in pituitary tumorigenesis and be involved in tumor cell proliferation and tumor progression. In addition, we provided here pathological evidence of a significant link between HMGA2 oncogene and let-7 miRNA. Loss and reduction of let-7 expression may contribute to elevated HMGA2 protein expression, resulting in pituitary tumorigenesis and progression. Future studies should be directed at detecting exact role of let-7 in each type of pituitary adenomas and uncovering the mechanism of let-7 aberrant expression in pituitary adenomas. Furthermore, let-7 miRNAs may be used as a novel anticancer agent in HMGA2-based therapy. To translate this finding into therapeutic strategy is a big challenge.
References
Asa SL, Ezzat S . The pathogenesis of pituitary tumours. Nat Rev Cancer 2002;2:836–849.
Ezzat S, Asa SL . Mechanisms of disease: the pathogenesis of pituitary tumors. Nat Clin Pract Endocrinol Metab 2006;2:220–230.
Reeves R, Nissen MS . The AT DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem 1990;265:8573–8582.
Thanos D, Maniatis T . The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 1992;27:777–789.
Thanos D, Du W, Maniatis T . The high mobility group protein HMG I(Y) is an essential structural component of a virus-inducible enhancer complex. Cold Spring Harb Symp Quant Biol 1993;58:73–81.
Zhou X, Benson KF, Ashar HR, et al. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995;376:771–774.
Chiappetta G, Avantaggiato V, Visconti R, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene 1996;13:2439–2446.
Sgarra R, Rustighi A, Tessari MA, et al. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett 2004;574:1–8.
Fusco A, Fedele M . Roles of HMGA proteins in cancer. Nat Rev Cancer 2007;7:899–910.
Fedele M, Battista S, Kenyon L, et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 2002;21:3190–3198.
Finelli P, Pierantoni GM, Giardino D, et al. The high mobility group A2 gene is amplified and overexpressed in human prolactinomas. Cancer Res 2002;62:2398–2405.
Pierantoni GM, Finelli P, Valtorta E, et al. High-mobility group A2 gene expression is frequently induced in non-functioning pituitary adenomas (NFPAs), even in the absence of chromosome 12 polysomy. Endocr Relat Cancer 2005;12:867–874.
Caldas C, Brenton JD . Sizing up miRNAs as cancer genes. Nat Med 2005;11:712–714.
Esquela-Kerscher A, Slack FJ . Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–269.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297.
Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide Let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901–906.
Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of Let-7 heterochronic regulatory RNA. Nature 2000;408:86–89.
Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–858.
Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635–647.
Mayr C, Hemann MT, Bartel DP . Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576–1579.
Hebert C, Norris K, Scheper MA, et al. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer 2007;6:5.
Wang T, Zhang X, Obijuru L, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer 2007;46:336–347.
Lee YS, Dutta A . The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007;21:1025–1030.
Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 2007;104:11400–11405.
Hardy J . Transsphenoidal microsurgical treatment of pituitary tumours. In: Linfoot J (ed). Recent Advances in the Diagnosis and Treatment of Pituitary Tumours. Raven Press: New York, 1979, pp 375–888.
Qian ZR, Sano T, Asa SL, et al. Cytoplasmic expression of fibroblast growth factor receptor-4 in human pituitary adenomas: relation to tumor type, size, proliferation, and invasiveness. J Clin Endocrinol Metab 2004;89:1904–1911.
Qian ZR, Sano T, Yoshimoto K, et al. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod Pathol 2007;20:1269–1277.
Livak KJ . ABI Prism 7700 Sequence Detection System. User Bulletin no. 2. PE Applied Biosystems, AB website, bulletin reference: 4303859B 777802-002 1997.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method 2001;5:402–408.
Fabjani G, Tong D, Wolf A, et al. HMGA2 is associated with invasiveness but not a suitable marker for the detection of circulating tumor cells in breast cancer. Oncol Rep 2005;14:737–741.
Motoyama K, Inoue H, Nakamura Y, et al. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res 2008;14:2334–2340.
Miyazawa J, Mitoro A, Kawashiri S, et al. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res 2004;64:2024–2029.
Thuault S, Valcourt U, Petersen M, et al. Transforming growth factor-β employs HMGA2 to elicit epithelial–mesenchymal transition. J Cell Biol 2006;174:175–183.
Pentimalli F, Dentice M, Fedele M, et al. Suppression of HMGA2 protein synthesis could be a tool for the therapy of well differentiated liposarcomas overexpressing HMGA2. Cancer Res 2003;63:7423–7427.
Simpson DJ, McNicol AM, Murray DC, et al. Molecular pathology shows p16 methylation in nonadenomatous pituitaries from patients with Cushing's disease. Clin Cancer Res 2004;10:1780–1788.
Simpson DJ, Hibberts NA, McNicol AM, et al. Loss of pRb expression in pituitary adenomas is associated with methylation of the RB1 CpG Island. Cancer Res 2000;60:1211–1216.
Qian ZR, Sano T, Yoshimoto K, et al. Inactivation of RASSF1A tumor suppressor gene by aberrant promoter hypermethylation in human pituitary adenomas. Lab Invest 2005;85:464–473.
Vallar L, Spada A, Giannattasio G . Altered Gs and adenylate cyclase activity in human GH-secreting pituitary adenomas. Nature 1987;330:566–568.
Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004.
Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753–3756.
Akao Y, Nakagawa Y, Naoe T . let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006;29:903–906.
Bottoni A, Zatelli MC, Ferracin M, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol 2007;210:370–377.
Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res 2007;67:1419–1423.
Peng Y, Laser J, Shi G, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res 2008;6:663–673.
Acknowledgements
We would like to express our appreciation to our patients. This study was partially funded by grants from the Ministry of Education, Science, and Technology, Japan (Knowledge Cluster Initiative, HI, MI).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Qian, Z., Asa, S., Siomi, H. et al. Overexpression of HMGA2 relates to reduction of the let-7 and its relationship to clinicopathological features in pituitary adenomas. Mod Pathol 22, 431–441 (2009). https://doi.org/10.1038/modpathol.2008.202
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.202
Keywords
This article is cited by
-
Genomics and Epigenomics of Pituitary Tumors: What Do Pathologists Need to Know?
Endocrine Pathology (2021)
-
Advancements in the study of miRNA regulation during the cell cycle in human pituitary adenomas
Journal of Neuro-Oncology (2017)
-
Increased expression of the microRNA 106b~25 cluster and its host gene MCM7 in corticotroph pituitary adenomas is associated with tumor invasion and Crooke’s cell morphology
Pituitary (2017)
-
Circulating miRNAs as biomarkers for endocrine disorders
Journal of Endocrinological Investigation (2016)
-
Gene expression in prolactinomas: a systematic review
Pituitary (2016)