Abstract
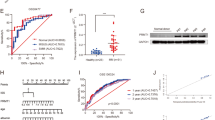
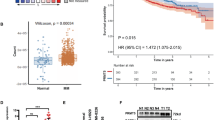
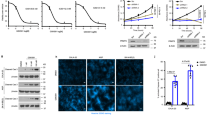
Arginine methyltransferases critically regulate cellular homeostasis by modulating the functional outcome of their substrates. The protein arginine methyltransferase 5 (PRMT5) is an enzyme involved in growth and survival pathways promoting tumorigenesis. However, little is known about the biologic function of PRMT5 and its therapeutic potential in multiple myeloma (MM). In the present study, we identified and validated PRMT5 as a new therapeutic target in MM. PRMT5 is overexpressed in patient MM cells and associated with decreased progression-free survival and overall survival. Either genetic knockdown or pharmacological inhibition of PRMT5 with the inhibitor EPZ015666 significantly inhibited growth of both cell lines and patient MM cells. Furthermore, PRMT5 inhibition abrogated NF-κB signaling. Interestingly, mass spectrometry identified a tripartite motif-containing protein 21 TRIM21 as a new PRMT5-partner, and we delineated a TRIM21-dependent mechanism of NF-κB inhibition. Importantly, oral administration of EPZ015666 significantly decreased MM growth in a humanized murine model of MM. These data both demonstrate the oncogenic role and prognostic relevance of PRMT5 in MM pathogenesis, and provide the rationale for novel therapies targeting PRMT5 to improve patient outcome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Anderson KC . Progress and paradigms in multiple myeloma. Clin Cancer Res 2016; 22: 5419–5427.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014; 5: 2997.
Bianchi G, Richardson PG, Anderson KC . Promising therapies in multiple myeloma. Blood 2015; 126: 300–310.
Ohguchi H, Hideshima T, Bhasin MK, Gorgun GT, Santo L, Cea M et al. The KDM3A-KLF2-IRF4 axis maintains myeloma cell survival. Nat Commun 2016; 7: 10258.
Amodio N, Stamato MA, Gulla AM, Morelli E, Romeo E, Raimondi L et al. Therapeutic targeting of miR-29b/HDAC4 epigenetic loop in multiple myeloma. Mol Cancer Ther 2016; 15: 1364–1375.
Hideshima T, Qi J, Paranal RM, Tang W, Greenberg E, West N et al. Discovery of selective small-molecule HDAC6 inhibitor for overcoming proteasome inhibitor resistance in multiple myeloma. Proc Natl Acad Sci USA 2016; 113: 13162–13167.
Amodio N, D'Aquila P, Passarino G, Tassone P, Bellizzi D . Epigenetic modifications in multiple myeloma: recent advances on the role of DNA and histone methylation. Expert Opin Ther Targets 2017; 21: 91–101.
Cheung N, Chan LC, Thompson A, Cleary ML, So CW . Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol 2007; 9: 1208–1215.
Copeland RA . Molecular pathways: protein methyltransferases in cancer. Clin Cancer Res 2013; 19: 6344–6350.
Stopa N, Krebs JE, Shechter D . The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 2015; 72: 2041–2059.
Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 2016; 351: 1214–1218.
Bedford MT, Clarke SG . Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009; 33: 1–13.
Yang Y, Bedford MT . Protein arginine methyltransferases and cancer. Nat Rev Cancer 2013; 13: 37–50.
Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S . Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J 2007; 26: 3558–3569.
Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov 2015; 5: 288–303.
Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun 2015; 6: 8419.
Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget 2015; 6: 22799–22811.
Alinari L, Mahasenan KV, Yan F, Karkhanis V, Chung JH, Smith EM et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 2015; 125: 2530–2543.
Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S . Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci 2011; 36: 633–641.
Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B et al. Arginine methylation regulates the p53 response. Nat Cell Biol 2008; 10: 1431–1439.
Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 2015; 11: 432–437.
Attal M, Lauwers-Cances V, Hulin C, Leleu X, Caillot D, Escoffre M et al. Lenalidomide, bortezomib, and dexamethasone with Transplantation for Myeloma. N Engl J Med 2017; 7: e554.
Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res 2007; 67: 2982–2989.
Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 2007; 109: 1692–1700.
Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J . Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia 2006; 20: 1288–1290.
Kawano Y, Roccaro AM, Azzi J, Ghobrial IM . Multiple Myeloma and the immune microenvironment. Curr Cancer Drug Targets 2017; 17: 806–818.
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P . The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1: 417–425.
Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem 2002; 277: 16639–16647.
Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 2007; 12: 115–130.
Wada K, Niida M, Tanaka M, Kamitani T . Ro52-mediated monoubiquitination of IKK{beta} down-regulates NF-{kappa}B signalling. J Biochem 2009; 146: 821–832.
Niida M, Tanaka M, Kamitani T . Downregulation of active IKK beta by Ro52-mediated autophagy. Mol Immunol 2010; 47: 2378–2387.
Kraft C, Peter M, Hofmann K . Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 2010; 12: 836–841.
Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell 2014; 30: 394–409.
Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E et al. Clinical significance of TP53 mutation in myeloma. Leukemia 2007; 21: 582–584.
Lode L, Eveillard M, Trichet V, Soussi T, Wuilleme S, Richebourg S et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 2010; 95: 1973–1976.
Hatakeyama S . TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 2017; 42: 297–311.
Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 2015; 210: 973–989.
Li S, Yang P, Tian E, Zhang H . Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol Cell 2013; 52: 421–433.
Stolz A, Ernst A, Dikic I . Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16: 495–501.
Calimeri T, Battista E, Conforti F, Neri P, Di Martino MT, Rossi M et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia 2011; 25: 707–711.
Tassone P, Neri P, Carrasco DR, Burger R, Goldmacher VS, Fram R et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood 2005; 106: 713–716.
Hideshima T, Chauhan D, Kiziltepe T, Ikeda H, Okawa Y, Podar K et al. Biologic sequelae of I{kappa}B kinase (IKK) inhibition in multiple myeloma: therapeutic implications. Blood 2009; 113: 5228.
Acknowledgements
This work has been supported by a grant from NIH P50−100707 (KCA, NCM), RO-1 CA050947 (KCA), and RO-1 CA 178264 (TH) and VA merit grant (I01BX001584) (NCM) and partially by the Italian Association for Cancer Research (AIRC) with ‘Special Program for Molecular Clinical Oncology–5 per mille’, 2010/15 and its Extension Program’ No. 9980, 2016/18 (PI: PT); and also by ‘Innovative Immunotherapeutic Treatments of Human Cancer’ Multi Unit Regional No. 16695 (cofinanced by AIRC and the CARICAL foundation), 2015/18 (PI: PT). KCA is an American Cancer Society Clinical Research Professor.
Author contributions
AG, TH and KCA designed research and wrote the manuscript; AG, GB, MF, TH, EM and NA performed the in vitro experiments and analyzed the data; MKS analyzed gene expression and RNA-seq data; RC performed IHC staining; JQ provided reagents and analytic tools; Y-TT and KCA provided MM patients samples; AG and EM designed, performed and analyzed in vivo experiments; NCM, PT and PT provided critical evaluation of experimental data and edited the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
NCM serves on advisory boards to Millennium, Celgene and Novartis. KCA serves on advisory boards Celgene, Millennium and Gilead Sciences and is a Scientific founder of OncoPep and C4 Therapeutics. All other authors declare no competing financial interests.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Gullà, A., Hideshima, T., Bianchi, G. et al. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia 32, 996–1002 (2018). https://doi.org/10.1038/leu.2017.334
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.334
This article is cited by
-
PRMT5 inhibition shows in vitro efficacy against H3K27M-altered diffuse midline glioma, but does not extend survival in vivo
Scientific Reports (2024)
-
PRMT1 methylation of WTAP promotes multiple myeloma tumorigenesis by activating oxidative phosphorylation via m6A modification of NDUFS6
Cell Death & Disease (2023)
-
Small molecule targeting of chromatin writers in cancer
Nature Chemical Biology (2022)
-
Structural insight into PRMT5 inhibitors through amalgamating pharmacophore-based virtual screening, ADME toxicity, and binding energy studies to identify new inhibitors by molecular docking
Structural Chemistry (2022)
-
Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition
Cell Death & Differentiation (2021)