Abstract
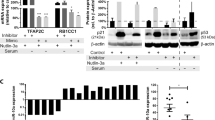
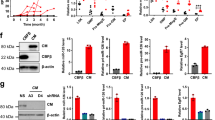
MYC is a major oncogenic driver of multiple myeloma (MM) and yet almost no therapeutic agents exist that target MYC in MM. Here we report that the let-7 biogenesis inhibitor LIN28B correlates with MYC expression in MM and is associated with adverse outcome. We also demonstrate that the LIN28B/let-7 axis modulates the expression of MYC, itself a let-7 target. Further, perturbation of the axis regulates the proliferation of MM cells in vivo in a xenograft tumor model. RNA-sequencing and gene set enrichment analyses of CRISPR-engineered cells further suggest that the LIN28/let-7 axis regulates MYC and cell cycle pathways in MM. We provide proof of principle for therapeutic regulation of MYC through let-7 with an LNA-GapmeR (locked nucleic acid-GapmeR) containing a let-7b mimic in vivo, demonstrating that high levels of let-7 expression repress tumor growth by regulating MYC expression. These findings reveal a novel mechanism of therapeutic targeting of MYC through the LIN28B/let-7 axis in MM that may impact other MYC-dependent cancers as well.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Howlader N, Noone AM, Yu M, Cronin KA . Use of imputed population-based cancer registry data as a method of accounting for missing information: application to estrogen receptor status for breast cancer. Am J Epidemiol 2012; 176: 347–356.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011; 25: 1026–1035.
Walker BA, Wardell CP, Murison A, Boyle EM, Begum DB, Dahir NM et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun 2015; 6: 6997.
Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014; 28: 1725–1735.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917.
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403: 901–906.
Roush S, Slack FJ . The let-7 family of microRNAs. Trends Cell Biol 2008; 18: 505–516.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67: 9762–9770.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64: 3753–3756.
Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 2007; 104: 11400–11405.
Emmrich S, Rasche M, Schoning J, Reimer C, Keihani S, Maroz A et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev 2014; 28: 858–874.
Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E et al. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood 2012; 119: 377–387.
Schulman BR, Esquela-Kerscher A, Slack FJ . Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn 2005; 234: 1046–1054.
Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 2007; 21: 415–426.
Viswanathan SR, Daley GQ, Gregory RI . Selective blockade of microRNA processing by Lin28. Science 2008; 320: 97–100.
Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009; 41: 843–848.
Feng C, Neumeister V, Ma W, Xu J, Lu L, Bordeaux J et al. Lin28 regulates HER2 and promotes malignancy through multiple mechanisms. Cell Cycle 2012; 11: 2486–2494.
King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK . LIN28B promotes colon cancer progression and metastasis. Cancer Res 2011; 71: 4260–4268.
Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006; 384: 51–61.
Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014; 26: 248–261.
Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012; 44: 1199–1206.
Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet 2012; 44: 1126–1130.
Urbach A, Yermalovich A, Zhang J, Spina CS, Zhu H, Perez-Atayde AR et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 2014; 28: 971–982.
Tu HC, Schwitalla S, Qian Z, LaPier GS, Yermalovich A, Ku YC et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev 2015; 29: 1074–1086.
International Myeloma Working G. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol 2003; 121: 749–757.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87.
Leleu X, Jia X, Runnels J, Ngo HT, Moreau AS, Farag M et al. The Akt pathway regulates survival and homing in Waldenstrom macroglobulinemia. Blood 2007; 110: 4417–4426.
Tomayko MM, Reynolds CP . Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 1989; 24: 148–154.
Shyh-Chang N, Daley GQ . Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013; 12: 395–406.
Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 2008; 105: 7004–7009.
Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J . Homozygous deletion scanning of the lung cancer genome at a 100- kb resolution. Genes Chromosomes Cancer 2007; 46: 1000–1010.
Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer 2008; 47: 810–818.
Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H . Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res 2007; 67: 10117–10122.
Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 2006; 9: 313–325.
Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 2010; 52: 1731–1740.
Wang J, Cao N, Yuan M, Cui H, Tang Y, Qin L et al. MicroRNA-125b/Lin28 pathway contributes to the mesendodermal fate decision of embryonic stem cells. Stem Cells Dev 2012; 21: 1524–1537.
Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008; 10: 987–993.
Segalla S, Pivetti S, Todoerti K, Chudzik MA, Giuliani EC, Lazzaro F et al. The ribonuclease DIS3 promotes let-7 miRNA maturation by degrading the pluripotency factor LIN28B mRNA. Nucleic Acids Res 2015; 43: 5182–5193.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014; 25: 91–101.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014; 5: 2997.
Acknowledgements
SM was supported by a grant from ARC Foundation. This work was supported by a grant from the National Cancer Institute (R01CA154648). Author Information: RNA-sequencing data have been deposited to the Gene Expression Omnibus under accession numbers GSE71100.
Author contributions
SM, JTP, GQD and IMG designed research; SM, AS, SVG, DH, MRR and YM performed in vitro research; SM, KS and MM performed in vivo research; JS processed RNA-sequencing data; SM, JTP, CR-L, XL, AMR, GQD and IMG analyzed data; SM, JTP, SVG, GQD and IMG wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Manier, S., Powers, J., Sacco, A. et al. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia 31, 853–860 (2017). https://doi.org/10.1038/leu.2016.296
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.296
This article is cited by
-
Research progress on the multi-omics and survival status of circulating tumor cells
Clinical and Experimental Medicine (2024)
-
In vitro and ex vivo anti-myeloma effects of nanocomposite As4S4/ZnS/Fe3O4
Scientific Reports (2022)
-
RNA-binding proteins in tumor progression
Journal of Hematology & Oncology (2020)
-
The effects of MicroRNA deregulation on pre-RNA processing network in multiple myeloma
Leukemia (2020)
-
Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target
Nature Communications (2020)