Abstract
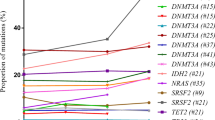
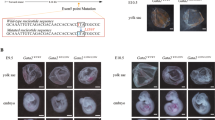
Acute erythroid leukemia (AEL), characterized by a predominant erythroid proliferation, is a subtype of acute myelogenous leukemia. The genetic basis of AEL remains poorly defined. Through whole-exome sequencing, we identified high frequencies of mutations in CEBPA (32.7%), GATA2 (22.4%), NPM1 (15.5%), SETBP1 (12.1%) and U2AF1 (12.1%). Structure prediction analysis revealed that most of the GATA2 mutations were located at the DNA-binding N-terminal zinc-finger near the DNA-binding interface, suggesting that mutations could result in at least partial inactivation of GATA2 protein. On co-transfection of a GATA-responsive reporter construct together with plasmids expressing either GATA2 wild-type or GATA2 ZF1 mutants (P304H, L321P and R330X) in 293T cells, we found a reduced transcriptional activation in cells transfected with GATA2 mutants. To determine whether reduced GATA2 function is involved in leukemogenesis of AEL, we transfected 32D cells with GATA2 mutants and evaluated the impact of GATA2 mutations on erythroid differentiation. Our data revealed an increased expression of erythroid-related antigens Ter-119, β-globin and βh1-globin, as well as increased hemoglobin positivity in 32D cells transfected with GATA2 mutants compared with control cells. Our results suggest that the decline of GATA2 resulting from mutations contributes to the erythroid commitment, differentiation and the development of AEL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 1985; 103: 620–625.
Arber DA, Brunning RD, Orazi A, Porwit A, Peterson L, Thiele J et al. Acute myeloid leukemia, not otherwise specified. In: Swerdlow SH, Campo E, Harris NL et al.(eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. International Agency for Research on Cancer (IARC): Lyon, France, 2008, pp 130–139..
Wells AW, Bown N, Reid MM, Hamilton PJ, Jackson GH, Taylor PR . Erythroleukaemia in the north of England: a population based study. J Clin Pathol 2001; 54: 608–612.
Hasserjian RP, Zuo Z, Garcia C, Tang G, Kasyan A, Luthra R et al. Acute erythroid leukemia: a reassessment using criteria refined in the 2008 WHO classification. Blood 2010; 115: 1985–1992.
The Cancer Genome Atlas Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–1066.
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3 A mutations in acute myeloid leukemia. N Engl J Med 2010; 363: 2424–2433.
Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 2013; 45: 1232–1237.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278.
Grossmann V, Bacher U, Haferlach C, Schnittger S, Pötzinger F, Weissmann S et al. Acute erythroid leukemia (AEL) can be separated into distinct prognostic subsets based on cytogenetic and molecular genetic characteristics. Leukemia 2013; 27: 1940–1943.
Cervera N, Carbuccia N, Garnier S, Guille A, Adélaïde J, Murati A et al. Molecular characterization of acute erythroid leukemia (M6-AML) using targeted next-generation sequencing. Leukemia 2015; 30: 966–970.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539.
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42: W252–W258.
Chen Y, Bates DL, Dey R, Chen PH, Machado AC, Laird-Offringa IA et al. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep 2012; 2: 1197–1206.
Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med 2004; 200: 871–882.
Tsai FY, Orkin SH . Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 1997; 89: 3636–3643.
Minegishi N, Ohta J, Yamagiwa H, Suzuki N, Kawauchi S, Zhou Y et al. The mouse GATA-2 gene is expressed in the para-aortic splanchnopleura and aorta-gonads and mesonephros region. Blood 1999; 93: 4196–4207.
Pasquet M, Bellanné-Chantelot C, Tavitian S, Prade N, Beaupain B, Larochelle O et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood 2013; 121: 822–829.
Trainor CD, Ghirlando R, Simpson MA . GATA zinc finger interactions modulate DNA binding and transactivation. J Biol Chem 2000; 275: 28157–28166.
Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994; 371: 221–226.
Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S . GATA switches as developmental drivers. J Biol Chem 2010; 285: 31087–31093.
Suzuki M, Kobayashi-Osaki M, Tsutsumi S, Pan X, Ohmori S, Takai J et al. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells 2013; 18: 921–933.
Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 1999; 93: 488–499.
Rodrigues NP, Boyd AS, Fugazza C, May GE, Guo Y, Tipping AJ et al. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood 2008; 112: 4862–4873.
Huang Z, Dore LC, Li Z, Orkin SH, Feng G, Lin S et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol 2009; 29: 5168–5180.
Zhang SJ, Ma LY, Huang QH, Li G, Gu BW, Gao XD et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci USA 2008; 105: 2076–2081.
Greif PA, Dufour A, Konstandin NP, Ksienzyk B, Zellmeier E, Tizazu B et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood 2012; 120: 395–403.
Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 2011; 43: 1012–1017.
Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood 2014; 123: 809–821.
Wlodarski MW, Hirabayashi S, Pastor V, Starý J, Hasle H, Masetti R et al. Prevalence, clinical characteristics and prognosis of GATA2-related myelodysplastic syndromes (MDS) in children and adolescents. Blood 2015; 127: 1387–1397.
Dickinson RE, Milne P, Jardine L, Zandi S, Swierczek SI, McGovern N et al. The evolution of cellular deficiency in GATA2 mutation. Blood 2014; 123: 863–874.
Acknowledgements
This work was supported by grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Natural Science Foundation of China (81570139 and 81270617), Jiangsu Provincial Special Program of Medical Science (BL2012005), Jiangsu Province’s Key Medical Center (ZX201102), Jiangsu Province Natural Science Foundation for Distinguished Young Scholars (BK2012006) and Jiangsu Province Natural Science Fund (BE2015639).
Author contributions
SC and DW were the principal investigators. NP, AS, QW, JY, WC, YX, LW, HY, HQ and LM performed most of the experiments. SC, CR, YS and DW wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Ping, N., Sun, A., Song, Y. et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia 31, 195–202 (2017). https://doi.org/10.1038/leu.2016.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.162
This article is cited by
-
Needle in a haystack or elephant in the room? Identifying germline predisposition syndromes in the setting of a new myeloid malignancy diagnosis
Leukemia (2023)
-
Breaking the spatial constraint between neighboring zinc fingers: a new germline mutation in GATA2 deficiency syndrome
Leukemia (2021)
-
Inherited GATA2 Deficiency Is Dominant by Haploinsufficiency and Displays Incomplete Clinical Penetrance
Journal of Clinical Immunology (2021)
-
Genomic subtyping and therapeutic targeting of acute erythroleukemia
Nature Genetics (2019)
-
Differential effects on gene transcription and hematopoietic differentiation correlate with GATA2 mutant disease phenotypes
Leukemia (2018)