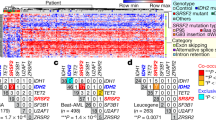
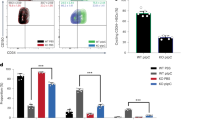
Abstract
Although the transforming potential of Hox genes is known for a long time, it is not precisely understood to which extent splicing is important for the leukemogenicity of this gene family. To test this for Hoxa9, we compared the leukemogenic potential of the wild-type Hoxa9, which undergoes natural splicing, with a full-length Hoxa9 construct, which was engineered to prevent natural splicing (Hoxa9FLim). Inability to undergo splicing significantly reduced in vivo leukemogenicity compared to Hoxa9-wild-typed. Importantly, Hoxa9FLim could compensate for the reduced oncogenicity by collaborating with the natural splice variant Hoxa9T, as co-expression of Hoxa9T and Hoxa9FLim induced acute myeloid leukemia (AML) after a comparable latency time as wild-type Hoxa9. Hoxa9T on its own induced AML after a similar latency as Hoxa9FLim, despite its inability to bind DNA. These data assign splicing a central task in Hox gene mediated leukemogenesis and suggest an important role of homeodomain-less splice variants in hematological neoplasms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hahn CN, Scott HS . Spliceosome mutations in hematopoietic malignancies. Nat Genet 2012; 44: 9–10.
Talavera D, Orozco M, de la Cruz X . Alternative splicing of transcription factors' genes: beyond the increase of proteome diversity. Comp Funct Genomics 2009; 2009: 905894.
Reed HC, Hoare T, Thomsen S, Weaver TA, White RA, Akam M et al. Alternative splicing modulates Ubx protein function in Drosophila melanogaster. Genetics 2010; 184: 745–758.
Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 1997; 89: 1922–1930.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531–537.
Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G . Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol 2001; 21: 224–234.
Rawat VP, Humphries RK, Buske C . Beyond HOX: the role of ParaHox genes in normal and malignant hematopoiesis. Blood 2012; 120: 519–527.
Breitinger C, Maethner E, Garcia-Cuellar MP, Slany RK . The homeodomain region controls the phenotype of HOX-induced murine leukemia. Blood 2012; 120: 4018–4027.
Fujimoto S, Araki K, Chisaka O, Araki M, Takagi K, Yamamura K . Analysis of the murine Hoxa-9 cDNA: an alternatively spliced transcript encodes a truncated protein lacking the homeodomain. Gene 1998; 209: 77–85.
Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest 2005; 115: 2159–2168.
Dintilhac A, Bihan R, Guerrier D, Deschamps S, Pellerin I . A conserved non-homeodomain Hoxa9 isoform interacting with CBP is co-expressed with the 'typical' Hoxa9 protein during embryogenesis. Gene Expr Patterns 2004; 4: 215–222.
Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell 2006; 10: 363–374.
Deshpande AJ, Rouhi A, Lin Y, Stadler C, Greif PA, Arseni N et al. The clathrin-binding domain of CALM and the OM-LZ domain of AF10 are sufficient to induce acute myeloid leukemia in mice. Leukemia 2011; 25: 1718–1727.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Williams GT, Farzaneh F . Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer 2012; 12: 84–88.
Bazeley PS, Shepelev V, Talebizadeh Z, Butler MG, Fedorova L, Filatov V et al. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 2008; 408: 172–179.
He M, Chen P, Arnovitz S, Li Y, Huang H, Neilly MB et al. Two isoforms of HOXA9 function differently but work synergistically in human MLL-rearranged leukemia. Blood Cells Mol Dis 2012; 49: 102–106.
Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK . Leukemogenic transformation by HOXA cluster genes. Blood 2010; 115: 2910–2918.
Shen WF, Detmer K, Simonitch-Eason TA, Lawrence HJ, Largman C . Alternative splicing of the HOX 2.2 homeobox gene in human hematopoietic cells and murine embryonic and adult tissues. Nucleic Acids Res 1991; 19: 539–545.
Schnabel CA, Jacobs Y, Cleary ML . HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 2000; 19: 608–616.
Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 2012; 119: 388–398.
Fernandez CC, Gudas LJ . The truncated Hoxa1 protein interacts with Hoxa1 and Pbx1 in stem cells. J Cell Biochem 2009; 106: 427–443.
Benson GV, Nguyen TH, Maas RL . The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol 1995; 15: 1591–1601.
Noro B, Culi J, McKay DJ, Zhang W, Mann RS . Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev 2006; 20: 1636–1650.
Acknowledgements
This work was supported by a grant of the Deutsche Krebshilfe (German Cancer Aid) (Project No 108438 to CB). We want to thank the Core Facility FACS, Ulm University and the team of the animal facility at Ulm University and at the Helmholtz Center Munich.
Author Contributions
CRS, NV, MAM, VPSR, KEE, AD, LB, BH and LQ-F performed research and KS, WH, KD, HD and MF-B provided patient samples and analyzed data. CB designed the research. CRS, NV and CB analyzed the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Stadler, C., Vegi, N., Mulaw, M. et al. The leukemogenicity of Hoxa9 depends on alternative splicing. Leukemia 28, 1838–1843 (2014). https://doi.org/10.1038/leu.2014.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.74