Abstract
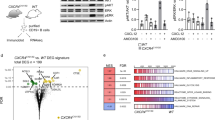
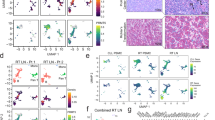
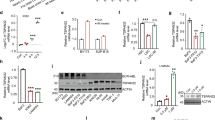
The gene encoding PTPROt (truncated isoform of protein tyrosine phosphatase receptor-type O) is methylated and suppressed in chronic lymphocytc leukemia (CLL). PTPROt exhibits in vitro tumor-suppressor characteristics through the regulation of B-cell receptor (BCR) signaling. Here we generated transgenic (Tg) mice with B-cell-specific expression of PTPROt. Although lymphocyte development is normal in these mice, crossing them with TCL1 Tg mouse model of CLL results in a survival advantage compared with the TCL1 Tg mice. Gene expression profiling of splenic B-lymphocytes before detectable signs of CLL followed by Ingenuity Pathway Analysis revealed that the most prominently regulated functions in TCL1 Tg vs non-transgenic (NTg) and TCL1 Tg vs PTPROt/TCL1 double Tg are the same and also biologically relevant to this study. Further, enhanced expression of the chemokine Ccl3, the oncogenic transcription factor Foxm1 and its targets in TCL1 Tg mice were significantly suppressed in the double Tg mice, suggesting a protective function of PTPROt against leukemogenesis. This study also showed that PTPROt-mediated regulation of Foxm1 involves activation of p53, a transcriptional repressor of Foxm1, which is facilitated through suppression of BCR signaling. These results establish the in vivo tumor-suppressive function of PTPROt and identify p53/Foxm1 axis as a key downstream effect of PTPROt-mediated suppression of BCR signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Wiggins RC, Wiggins JE, Goyal M, Wharram BL, Thomas PE . Molecular cloning of cDNAs encoding human GLEPP1, a membrane protein tyrosine phosphatase: characterization of the GLEPP1 protein distribution in human kidney and assignment of the GLEPP1 gene to human chromosome 12p12-p13. Genomics 1995; 27: 174–181.
Wharram BL, Goyal M, Gillespie PJ, Wiggins JE, Kershaw DB, Holzman LB et al. Altered podocyte structure in GLEPP1 (Ptpro)-deficient mice associated with hypertension and low glomerular filtration rate. J Clin Invest 2000; 106: 1281–1290.
Beltran PJ, Bixby JL, Masters BA . Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. J Comp Neurol 2003; 456: 384–395.
Aguiar RC, Yakushijin Y, Kharbanda S, Tiwari S, Freeman GJ, Shipp MA . PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood 1999; 94: 2403–2413.
Hsu SH, Motiwala T, Roy S, Claus R, Mustafa M, Plass C et al. Methylation of the PTPRO gene in human hepatocellular carcinoma and identification of VCP as its substrate. J Cell Biochem 2013; 114: 1810–1818.
Ramaswamy B, Majumder S, Roy S, Ghoshal K, Kutay H, Datta J et al. Estrogen-mediated suppression of the gene encoding protein tyrosine phosphatase PTPRO in human breast cancer: mechanism and role in tamoxifen sensitivity. Mol Endocrinol 2009; 23: 176–187.
Motiwala T, Majumder S, Kutay H, Smith DS, Neuberg DS, Lucas DM et al. Methylation and silencing of protein tyrosine phosphatase receptor type O in chronic lymphocytic leukemia. Clin Cancer Res 2007; 13: 3174–3181.
Motiwala T, Kutay H, Ghoshal K, Bai S, Seimiya H, Tsuruo T et al. Protein tyrosine phosphatase receptor-type O (PTPRO) exhibits characteristics of a candidate tumor suppressor in human lung cancer. Proc Natl Acad Sci USA 2004; 101: 13844–13849.
Motiwala T, Ghoshal K, Das A, Majumder S, Weichenhan D, Wu YZ et al. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene 2003; 22: 6319–6331.
Jacob ST, Motiwala T . Epigenetic regulation of protein tyrosine phosphatases: potential molecular targets for cancer therapy. Cancer Gene Ther 2005; 12: 665–672.
Motiwala T, Majumder S, Ghoshal K, Kutay H, Datta J, Roy S et al. PTPROt inactivates the oncogenic fusion protein BCR/ABL and suppresses transformation of K562 cells. J Biol Chem 2009; 284: 455–464.
Shintani T, Ihara M, Sakuta H, Takahashi H, Watakabe I, Noda M . Eph receptors are negatively controlled by protein tyrosine phosphatase receptor type O. Nat Neurosci 2006; 9: 761–769.
Chen L, Juszczynski P, Takeyama K, Aguiar RC, Shipp MA . Protein tyrosine phosphatase receptor-type O truncated (PTPROt) regulates SYK phosphorylation, proximal B-cell-receptor signaling, and cellular proliferation. Blood 2006; 108: 3428–3433.
Motiwala T, Datta J, Kutay H, Roy S, Jacob ST . Lyn kinase and ZAP70 are substrates of PTPROt in B-cells: Lyn inactivation by PTPROt sensitizes leukemia cells to VEGF-R inhibitor pazopanib. J Cell Biochem 2010; 110: 846–856.
Huang YT, Li FF, Ke C, Li Z, Li ZT, Zou XF et al. PTPRO promoter methylation is predictive of poorer outcome for HER2-positive breast cancer: indication for personalized therapy. J Transl Med 2013; 11: 245.
You YJ, Chen YP, Zheng XX, Meltzer SJ, Zhang H . Aberrant methylation of the PTPRO gene in peripheral blood as a potential biomarker in esophageal squamous cell carcinoma patients. Cancer Lett 2012; 315: 138–144.
American Cancer Society: Cancer Facts and Figures. American Cancer Society, 2012. Available at http://www.cancer.org/research/cancerfactsstatistics/.
Motiwala T, Zanesi N, Datta J, Roy S, Kutay H, Checovich AM et al. AP-1 elements and TCL1 protein regulate expression of the gene encoding protein tyrosine phosphatase PTPROt in leukemia. Blood 2011; 118: 6132–6140.
Pekarsky Y, Zanesi N, Croce CM . Molecular basis of CLL. Semin Cancer Biol 2010; 20: 370–376.
Zanesi N, Balatti V, Bottoni A, Croce CM, Pekarsky Y . Novel insights in molecular mechanisms of CLL. Curr Pharm Des 2012; 18: 3363–3372.
Herling M, Patel KA, Khalili J, Schlette E, Kobayashi R, Medeiros LJ et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia 2006; 20: 280–285.
Zanesi N, Aqeilan R, Drusco A, Kaou M, Sevignani C, Costinean S et al. Effect of rapamycin on mouse chronic lymphocytic leukemia and the development of nonhematopoietic malignancies in Emu-TCL1 transgenic mice. Cancer Res 2006; 66: 915–920.
Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA 2002; 99: 6955–6960.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol 2010; 6: 377.
Enzler T, Kater AP, Zhang W, Widhopf GF 2nd, Chuang HY, Lee J et al. Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood 2009; 114: 4469–4476.
Burger JA, Quiroga MP, Hartmann E, Burkle A, Wierda WG, Keating MJ et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood 2009; 113: 3050–3058.
Sivina M, Hartmann E, Kipps TJ, Rassenti L, Krupnik D, Lerner S et al. CCL3 (MIP-1alpha) plasma levels and the risk for disease progression in chronic lymphocytic leukemia. Blood 2011; 117: 1662–1669.
Barsotti AM, Prives C . Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 2009; 28: 4295–4305.
Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther 2011; 10: 1046–1058.
Dai C, Gu W . p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 2010; 16: 528–536.
Schall TJ, Bacon K, Camp RD, Kaspari JW, Goeddel DV . Human macrophage inflammatory protein alpha (MIP-1 alpha) and MIP-1 beta chemokines attract distinct populations of lymphocytes. J Exp Med 1993; 177: 1821–1826.
Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R et al. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood 2011; 117: 5463–5472.
Liu J, Chen G, Feng L, Zhang W, Pelicano H, Wang F et al. Loss of p53 and altered miR15-a/16-1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53(−/−) mouse model and primary human leukemia cells. Leukemia 2014; 28: 118–128.
Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 2008; 112: 3322–3329.
Acknowledgements
We thank the Ohio State University Comprehensive Cancer Center Microarray, Analytical Cytometry and Genetically Engineered Mouse Modeling Shared Resources for technical assistance. We thank Kalpana Ghoshal, Sarmila Majumder and Jharna Datta for valuable discussions; Satavisha Roy for experimental assistance; and Alexey Efanov for help with flow cytometry. This work was supported, in part, by the NIH grants CA101956 and CA086978 from the National Cancer Institute (to STJ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Motiwala, T., Kutay, H., Zanesi, N. et al. PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model. Leukemia 29, 1350–1359 (2015). https://doi.org/10.1038/leu.2014.341
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.341
This article is cited by
-
Protein tyrosine phosphatase PTPRO represses lung adenocarcinoma progression by inducing mitochondria-dependent apoptosis and restraining tumor metastasis
Cell Death & Disease (2024)
-
Upregulation of FOXM1 leads to diminished drug sensitivity in myeloma
BMC Cancer (2018)
-
Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia
Oncogene (2018)
-
The PTPROt tyrosine phosphatase functions as an obligate haploinsufficient tumor suppressor in vivo in B-cell chronic lymphocytic leukemia
Oncogene (2017)
-
FOXM1 is a therapeutic target for high-risk multiple myeloma
Leukemia (2016)