Abstract
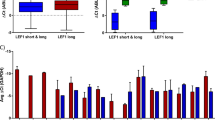
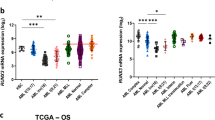
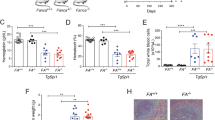
In leukemogenesis, Notch signaling can be up and downregulated in a context-dependent manner. The transcription factor hairy and enhancer of split-1 (Hes1) is well-characterized as a downstream target of Notch signaling. Hes1 encodes a basic helix–loop–helix-type protein, and represses target gene expression. Here, we report that deletion of the Hes1 gene in mice promotes acute myeloid leukemia (AML) development induced by the MLL–AF9 fusion protein. We then found that Hes1 directly bound to the promoter region of the FMS-like tyrosine kinase 3 (FLT3) gene and downregulated the promoter activity. FLT3 was consequently upregulated in MLL–AF9-expressing immortalized and leukemia cells with a Hes1- or RBPJ-null background. MLL–AF9-expressing Hes1-null AML cells showed enhanced proliferation and ERK phosphorylation following FLT3 ligand stimulation. FLT3 inhibition efficiently abrogated proliferation of MLL–AF9-induced Hes1-null AML cells. Furthermore, an agonistic anti-Notch2 antibody induced apoptosis of MLL–AF9-induced AML cells in a Hes1-wild type but not a Hes1-null background. We also accessed two independent databases containing messenger RNA (mRNA) expression profiles and found that the expression level of FLT3 mRNA was negatively correlated with those of HES1 in patient AML samples. These observations demonstrate that Hes1 mediates tumor suppressive roles of Notch signaling in AML development, probably by downregulating FLT3 expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Artavanis-Tsakonas S, Rand MD, Lake RJ . Notch signaling: cell fate control and signal integration in development. Science 1999; 284: 770–776.
De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 1999; 398: 518–522.
Kageyama R, Ohtsuka T, Tomita K . The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol Cells 2000; 10: 1–7.
Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol 2001; 21: 7761–7774.
Bigas A, Espinosa L . Hematopoietic stem cells: to be or Notch to be. Blood 2012; 119: 3226–3235.
Grbavec D, Stifani S . Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun 1996; 223: 701–705.
Ito T, Udaka N, Okudela K, Yazawa T, Kitamura H . Mechanisms of neuroendocrine differentiation in pulmonary neuroendocrine cells and small cell carcinoma. Endocr Pathol 2003; 14: 133–139.
Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 2000; 127: 3913–3921.
Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Suzuki T, Chiba S . Notch signaling in hematopoietic stem cells. Int J Hematol 2005; 82: 285–294.
Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999; 10: 547–558.
Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105.
Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012; 119: 1963–1971.
Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci 2009; 100: 920–926.
Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med 2012; 209: 1553–1565.
Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 2011; 17: 1646–1651.
Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet 2003; 33: 416–421.
Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB et al. Chronic DLL4 blockade induces vascular neoplasms. Nature 2010; 463: E6–E7.
Kannan S, Fang W, Song G, Mullighan CG, Hammitt R, McMurray J et al. Notch/HES1-mediated PARP1 activation: a cell type-specific mechanism for tumor suppression. Blood 2011; 117: 2891–2900.
Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 2010; 115: 2872–2881.
Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010; 466: 765–768.
Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 2011; 473: 230–233.
Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 2013; 210: 301–319.
Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med 2013; 210: 321–337.
Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol 2002; 14: 637–645.
Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol 2002; 3: 443–450.
Guiu J, Shimizu R, D'Altri T, Fraser ST, Hatakeyama J, Bresnick EH et al. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J Exp Med 2013; 210: 71–84.
Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F et al. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 1996; 16: 723–734.
Somervaille TC, Cleary ML . Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 2006; 10: 257–268.
Sang L, Coller HA, Roberts JM . Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 2008; 321: 1095–1100.
McLarren KW, Theriault FM, Stifani S . Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J Biol Chem 2001; 276: 1578–1584.
Kang HJ, Lee JW, Kho SH, Kim MJ, Seo YJ, Kim H et al. High transcript level of FLT3 associated with high risk of relapse in pediatric acute myeloid leukemia. J Korean Med Sci 2010; 25: 841–845.
Ozeki K, Kiyoi H, Hirose Y, Iwai M, Ninomiya M, Kodera Y et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood 2004; 103: 1901–1908.
Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene 2000; 19: 624–631.
Komeno Y, Kurokawa M, Imai Y, Takeshita M, Matsumura T, Kubo K et al. Identification of Ki23819, a highly potent inhibitor of kinase activity of mutant FLT3 receptor tyrosine kinase. Leukemia 2005; 19: 930–935.
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004; 350: 1617–1628.
Pigazzi M, Masetti R, Bresolin S, Beghin A, Di Meglio A, Gelain S et al. MLL partner genes drive distinct gene expression profiles and genomic alterations in pediatric acute myeloid leukemia: an AIEOP study. Leukemia 2011; 25: 560–563.
Moriyama Y, Sekine C, Koyanagi A, Koyama N, Ogata H, Chiba S et al. Delta-like 1 is essential for the maintenance of marginal zone B cells in normal mice but not in autoimmune mice. Int Immunol 2008; 20: 763–773.
Paganin M, Ferrando A . Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev 2011; 25: 83–90.
Samon JB, Castillo-Martin M, Hadler M, Ambesi-Impiobato A, Paietta E, Racevskis J et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther 2012; 11: 1565–1575.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 2006; 20: 2096–2109.
Wendorff AA, Koch U, Wunderlich FT, Wirth S, Dubey C, Brüning JC et al. Hes1 is a critical but context-dependent mediator of canonical notch signaling in lymphocyte development and transformation. Immunity 2010; 33: 671–684.
Espinosa L, Cathelin S, D'Altri T, Trimarchi T, Statnikov A, Guiu J et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell 2010; 18: 268–281.
Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210.
Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood 2002; 100: 4372–4380.
Riccioni R, Pelosi E, Riti V, Castelli G, Lo-Coco F, Testa U . Immunophenotypic features of acute myeloid leukaemia patients exhibiting high FLT3 expression not associated with mutations. Br J Haematol 2011; 153: 33–42.
Kuchenbauer F, Kern W, Schoch C, Kohlmann A, Hiddemann W, Haferlach T et al. Detailed analysis of FLT3 expression levels in acute myeloid leukemia. Haematologica 2005; 90: 1617–1625.
Kindler T, Lipka DB, Fischer T . FLT3 as a therapeutic target in AML: still challenging after all these years. Blood 2010; 116: 5089–5102.
Wang QF, Wu G, Mi S, He F, Wu J, Dong J et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood 2011; 117: 6895–6905.
Acknowledgements
We thank Drs T Machino and T Enami (University of Tsukuba) for discussion; Drs A Yokoyama (Kyoto University), H Nakauchi (University of Tokyo/Stanford University), and, M Onodera (National Research Institute for Child Health and Development) for vectors. We also thank T Takahashi for mouse experiments. We are also grateful to Kyowa Hakko Kirin Co., Ltd. for KRN383. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (25860778 to TK; 25461407 to MS-Y; and 25112703, 24390241, 23118503 and 22130002 to SC) and supported by the Sagawa Cancer Foundation, the Naito Foundation, the Kato Memorial Bioscience Foundation and the YASUDA Medical Foundation to MS-Y.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Kato, T., Sakata-Yanagimoto, M., Nishikii, H. et al. Hes1 suppresses acute myeloid leukemia development through FLT3 repression. Leukemia 29, 576–585 (2015). https://doi.org/10.1038/leu.2014.281
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.281
This article is cited by
-
Notch activator cyclopiazonic acid induces apoptosis in HL-60 cells through calcineurin activation
The Journal of Antibiotics (2024)
-
The fatty acid elongase Elovl6 is crucial for hematopoietic stem cell engraftment and leukemia propagation
Leukemia (2023)
-
Combined inhibition of Notch and FLT3 produces synergistic cytotoxic effects in FLT3/ITD+ acute myeloid leukemia
Signal Transduction and Targeted Therapy (2020)
-
Meis2 as a critical player in MN1-induced leukemia
Blood Cancer Journal (2017)
-
MiRNA182 regulates percentage of myeloid and erythroid cells in chronic myeloid leukemia
Cell Death & Disease (2017)