Abstract
Parkinson’s disease (PD) is the second leading neurodegenerative disease, and is known to be induced by environmental factors or genetic mutations. Among the verified genetic mutations of PD, Parkin, isolated from the PARK2 locus, shows an autosomal recessive inheritance pattern and is known to be an E3 ligase. However, the physiological target of Parkin and the molecular mechanism of Parkin-deficiency-induced PD have not been clearly demonstrated until now. It has recently been proposed that inflammation, suggesting as a causal factor for PD, is enhanced by Parkin deficiency. Thus, we examined the relationship between inflammation-related factors and Parkin. Here, we provide the evidence that Parkin suppresses inflammation and cytokine-induced cell death by promoting the proteasomal degradation of TRAF2/6 (TNF-α receptor-associated factor 2/6). Overexpression of Parkin can reduce the half-lives of TRAF2 and TRAF6, whereas si-Parkin can extend them. However, mutant Parkins did not alter the expression of TRAF2/6. Thus, loss of Parkin enhances sensitivity to TNF-α- or IL-1β-induced JNK activation and NF-κB activation. Indeed, si-Parkin-induced apoptosis is suppressed by the knockdown of TRAF6 or TRAF2. We also observed elevated expression levels of TRAF6 and a reduction of IκB in an 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced PD mouse model. Moreover, elevated expression levels or aggregation of TRAF6 were detected in approximately half of the human PD tissues (7/15 cases) and 2 cases, respectively. In addition, TRAF6 and Parkin expression levels show a reverse relationship in human PD tissues. Our results strongly suggest that the reduction of Parkin or overexpression of TRAF2/6 by chronic inflammation would be the reason for occurrence of PD.
Similar content being viewed by others
Main
Parkinson’s disease (PD) is a leading neurodegenerative movement disorder, and it is caused by the elimination of dopaminergic neurons in the substantia nigra (SN).1 Slow movement, resting tremors, postural instability, and muscle rigidity are well-known clinical symptoms of PD.2 Because the incidence of PD increases in an age-dependent manner, accumulated damages are also suggested as the reason of PD.3 In addition, chronic inflammation has been suggested as a risk factor of PD, similar to other neurodegenerative diseases, including Alzheimer’s disease and amyotrophic lateral sclerosis.4, 5 However, neither detailed molecular relevance nor a mechanism connecting inflammation and neurodegenerative diseases have been clearly demonstrated until now. With respect to the molecular pathological features of PD, the Lewy body (LB), which is mainly composed of α-synuclein, has been proposed as a molecular marker and causal alteration of PD.3, 6 However, it is not clear that LB is a causal factor of PD or the result of PD. Moreover, some subtypes of PD, such as Parkin-deficient juvenile-type PD, do not show the LB,7, 8 suggesting that LB may not be responsible for all types of PD.
Through genetic studies, putative genetic loci have been proposed and Parkin has been isolated from the PARK2 locus.2, 9 In contrast to other PD-related genes, such as α-synuclein or LRRK2, loss of Parkin function is related to the occurrence of PD.2, 7, 9 However, patients whose PD is induced by Parkin deficiency do not show the LB, indicating that the, loss of functional Parkin does not promote the formation of LB despite the activity of E3 ligase.9 The results of intensive studies to verify the target of the Parkin E3 ligase have suggested several protein targets for Parkin, including CDCrel-1, Pael-R, AIMP2, etc.10, 11, 12 However, these target proteins did not provide a sufficient explanation for Parkin-deficiency-induced neuronal cell death. Thus, the physio-pathological target of Parkin has been unknown until now. Recently, based on the frequent deletion or mutation in human colon cancer, Parkin has been suggested as a haploinsufficient tumor suppressor gene.13, 14 As chronic inflammation is tightly related with colon cancer as well as PD,15, 16 we examined the biological role of Parkin in the cytokine-induced inflammation response.
In this study, we reveal that Parkin promotes the proteasomal degradation of TRAF2/6 and protects inflammatory signaling. Indeed, Parkin expression is inversely related to TRAF6 expression. Moreover, mutant Parkin promotes TRAF6 aggregation in the cytosol. These results strongly suggest that TRAF2/6 is a physiological target of Parkin, and they provide an explanation for why loss of Parkin is detected in neurodegenerative diseases and cancer.
MATERIALS AND METHODS
Cell Lines and Reagents
The cell lines used in this study were maintained in RPMI-1640 (HCT116, DLD-1) and DMEM (293, SH-SY5Y, and N2A) supplied with 10% FBS in a 37 °C humidity chamber. Parkin-deficient lymphocytes were provided by the Coriell Institute for Medical Research. Antibodies against p-JNK, TRAF2, and Lamin were obtained from Cell Signaling (Cell Signaling Technology, Beverly, MA, USA). Antibodies for Parkin, GST, actin, ubiquitin, and TRAF6 were supplied by Santa Cruz Biotech (CA, USA). TNF-α and IL-1β were obtained from Peprotech. N-acetyl-cysteine (NAC) was purchased from Sigma (Sigma-Aldrich, St Louis, MO, USA). The luciferase assay kit was provided by Promega. Other general chemicals were obtained from Sigma (Sigma-Aldrich). To measure the NF-κB activity and translocation, SH-SY5Y cells were transfected with wild-type Parkin or si-parkin. After 24 h of incubation, the cells were treated with 10 mM NAC 1 h before the treatment of cytokines in serum-free media. For western blot, NAC was treated in the cells with IL-1β or TNF-α.
Vectors and Transfection
Mammalian expression vectors (Parkin, TRAF6 and mutant Parkin) were obtained from Addgene. The NF-κB luciferase vector was kindly provided by Chung HY (Pusan National Univ). The construct used for si-RNA against Parkin was generated as previously described.9 The transfections were performed following the manufacturer’s protocol. In brief, 1.5 μg of plasmid was mixed with 3 μl of Jetpei reagent (Polyfect) in a 150 mM NaCl solution. After 30 min incubation, the mixture was added to a monolayer of cells. After 3 h of incubation in serum-free conditions, the medium was replaced with complete medium. After 24 h, the cells were treated with TNF-α or IL-1β or harvested for experiments. To obtain the Parkin stable knock-out cell lines, sh-parkin was transfected with SHSY-5Y cells. Multiple constructs of PARK2 shRNA lentivial plasmids were purchased from Sigma. Multiple constructs to stably knock-out Parkin gene was used following shRNA sequence: 3′UTR, 5′-CCGGCGTGAACATAACTGAGGGCATCTCGAGATGCCCTCAGTTATGTTCACGTTTTT-3′; sh-parkin #1, 5′-CCGGCGCAACAAATAGTCGGAACATCTCGAGATGTTCCGACTATTTGTTGCGTTTTT-3′; sh-parkin #2, 5′-CCGGCGTGATTTGCTTAGACTGTTTCTCGAGAAACAGTCTAAGCAAATCACGTTTTT-3′; sh-parkin #3, 5′-CCGGCTCCAAAGAAACCATCAAGAACTCGAGTTCTTGATGGTTTCTTTGGAGTTTTT-3′; sh-parkin #4, 5′-CCGGCTTAGACTGTTTCCACTTATACTCGAGTATAAGTGGAAACAGTCTAAGTTTTT-3′.
Construction of TRAF6 Mutant
To construct the Traf6 alleles K91R, K96R, and K104R, a fragment of pGEX-TEV-Traf6 consisting of residues 50–159 was amplified by overlapping PCR using a common forward primer (5′-GGGGAATTCTAATGAGTCTCTTAAACTGTGAG-3′) and common reverse primer (5′-GGGAAGCTTCTACACCCCCGCATCAGTACTTCG-3′). We also used forward mutation primers K91R- 5′-GCGGCCATAGGTTCTGCAGAGCCTGCATC-3′, K96R- 5′-AGGTTCTGCAAAGCCTGCATCATAAGATCAATAAGGGATG-3′, and K104R- 5′-AAGGGATGCAGGTCACAGATGTCCAGTTGACAATG-3′, and reverse mutation primers K91R- 5′-GATGCAGGCTCTGCAGAACCTATGGCCGC-3′, K96R- 5′-CATCCCTTATTGATCTTATGATGCAGGCTTTGCAGAACCT-3′, and K104R- 5′-CATTGTCAACTGGACATCTGTGACCTGCATCCCTT-3′. The PCR products were inserted into the EcoRI and HindIII sites of the pGEX-TEV vector, respectively. The ligated vectors were transformed into XL-1 blue cells. Nucleotide sequencing was performed after plasmid preparation.
Protein Expression and Purification
Mutant TRAF6 proteins were expressed from E. coli BL21 cells (DE3) in 500 ml LB medium containing ampicillin (50 mg/ml) at 37 °C until the OD600 reached 0.6-0.8. Protein expression was induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 30 °C. After induction for 4 h, the cells were harvested by centrifugation at 5000 r.p.m. for 10 min at 4 °C. The harvested cells were disrupted by sonication in a lysis buffer consisting of 20 mM Tris, pH 8.0, 150 mM NaCl, and 2 mM β-mercaptoethanol. The soluble lysate was centrifuged at 13 000 r.p.m. for 30 min to remove any insoluble materials. The supernatant was mixed with a glutathione affinity resin that had been pre-incubated with lysis buffer, and the mixture was then stirred for 1 h at 4 °C. After the slurry was loaded onto the column, unbound proteins were washed off with lysis buffer.
MTT Assay
To measure cell viability, cells were transfected with either vectors or si-RNA for 24 h. After washing, cells were incubated with CHX for 4 h and TNF-α or IL-1β was added. For the MTT assay, cells were incubated with 0.5 mg/ml of MTT solution for 4 h at 37 °C. After the excess solution was removed, the precipitated materials were dissolved in 200 μl DMSO and quantified by measuring their absorbance at 540 nm.
Analysis of Protein Half-Life
To measure the half-life of TRAF2 and TRAF6, cells were transfected with Parkin or si-Parkin for 24 h. After the media was replaced, the cells were incubated with CHX to block de novo protein synthesis. After harvest, expression levels of TRAF2 and TRAF6 were determined by western blot analysis. More detailed protocols are available in our previous publication.17
Western Blot Analysis and GST-Pull Down Assay
To monitor the expression of proteins and in vitro binding, we performed the general western blot and GST-pull down assays.18
Luciferase Assay
To address NF-κB activity, an NF-κB-luc vector was transfected into 293 cells for 24 h. After washing, cells were stimulated by TNF-α or IL-1β for 4 h. After washing with wash buffer (Promega), cells were lysed with lysis buffer. The luciferase activity was determined using luminometer measurements.
Immunofluorescence Staining
The 293 cells were transfected with a Parkin or si-Parkin vector for 24 h and fixed with 100% Me-OH. After blocking in PBS containing 1% BSA for 2 h, the cells were incubated overnight with a TRAF6 (1:100) or Parkin (HA) antibody. TRAF6 was visualized with a FITC-conjugated secondary antibody and Parkin with a rhodamine antibody. DAPI was used for DNA staining.
Ubiquitination Analysis
The 293 cells were transfected with wild-type, mutant Parkin or si-Parkin for 24 h. After cells were washed, they were treated with ALLN for 3 h to block protein degradation. The cells were harvested, and the cell lysate was incubated sequentially with a TRAF2 antibody and protein A/G agarose beads for 2 h. After precipitation by centrifugation, the precipitated materials were subjected to WB analysis with an anti-Ub antibody. To determine the Ub-conjugated site on TRAF6, we generated three types of GST-fused mutant TRAF6-N-terminal proteins (K91R, K96R, and K104R). Recombinant proteins were incubated with Parkin and HA-Ub-transfected 293 cell lysates for 1 h. After washing, Ub-conjugated TRAF6 was analyzed by WB.
Mouse Brain Tissue Staining
For double-labeling IHC, the sections were blocked with TBS/0.1% Triton X-100/3% goat serum (Gibco, Grand Island, NY, USA) and incubated with primary antibodies against TRAF6 (mouse monoclonal, 1:100) and the astrocyte marker GFAP (rabbit polyclonal, 1:500, Sigma-Aldrich) at 4°C overnight. The brain sections were then washed with TBS and incubated for 3 h in the presence of anti-mouse IgG labeled with Alexa Fluor-568 and anti-rabbit IgG labeled with Alexa Fluor-488. Images were acquired using an FV10i FLUOVIEW Confocal Microscope (Olympus, Tokyo).
Human Tissue Analysis
Human PD and normal tissues were obtained from the Victorian Brain Bank network as paraffin sections. After a de-paraffining procedure using Xylene–Ethanol gradients, the tissues were incubated with TRAF6 or Parkin Antibodies (1:100) overnight. After the tissues were washed twice with PBS, HRP-conjugated anti-mouse antibody was added. After a 4 h reaction, washed slides were reacted with a DAB solution (Vector) for 5 min. After washing and counterstaining with hematoxylin, the slides were mounted.
Animals and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Injection
ICR mice were used for this study. A control (PBS-injected) and an MPTP-treated group were each made up of seven mice. The animals were maintained in a temperature-controlled room under a 12:12-h light/dark cycle with ad libitum access to food and water for 1 week before the experiment. The mice were injected intraperitoneally (i.p.) five times with 30 mg/kg/day MPTP-HCl or the corresponding volume of saline.
Statistical Analysis
To obtain the statistical significance, we performed the Student’s t-test.
RNA Isolation and RT-PCR
For RT-PCR, total cellular RNA was extracted using a Qiagen RNA extraction kit. After measurement of the RNA concentration, 1 μg of total RNA was reverse transcribed to cDNA using MMLV-RT (Invitrogen) and random hexamers. RT-PCR was performed with the primers specific to the TRAF2 (5′-TTCCCCTTAACTTGTGACGGC-3′ and 5′-CAATCTTGTCTTGGTCCAGCC-3′) and TRAF6 (5′-TACGAGAAGCAGTGCAAACGC-3′ and 5′-ATTTTTGGAAGGGACGCTGGC-3′) or GAPDH (5′-ATCTTCCAGGAGCGAGATCCC-3′ and 5′-AGTGAGCTTCCC GTTCAGCTC-3′).
RESULTS
Parkin Blocks Cytokine Signaling
Because elevated expression of TNF-α and IL-1β has been reported in PD patients,4, 19, 20, 21 we monitored the effect of Parkin on JNK activation, which is a well-established marker for the activation of these cytokine signaling pathways. Compared with the empty vector (EV)-transfected cells, induction of phospho-JNK (p-JNK) was suppressed in the Parkin-transfected cells (Figure 1a). We also observed delayed or completely blocked JNK activation in response to cytokines in Parkin-transfected cells over time (Supplementary Material, Supplementary Figure S1A and B). In neuronal cells, we confirmed the inhibitory role of Parkin on JNK activation (Figure 1b). Because these cytokines can also activate NF-κB signaling, we assayed the effect of Parkin on NF-κB signaling and found that the degradation of IκB was blocked by Parkin transfection (Supplementary Figure S1C and D). To confirm this result, we measured the expression of IκB in Parkin-deficient lymphocytes and observed that its levels were reduced when compared with Jurkat (Figure 1c). Parkin was also able to suppress the transcriptional activity of NF-κB (Figure 1d). In contrast, si-RNA knockdown of Parkin (Supplementary Figure S1E) enhanced the activation of JNK (Figure 1e) and the transcriptional function of NF-κB (Supplementary Figure S1F). We obtained similar results from human neuronal cells, where we found that Parkin could suppress the transcriptional activity of NF-κB and si-Parkin enhanced it (Figure 1f). Therefore, we next examined the translocation of NF-κB because impairment of IκB degradation may lead to defects in NF-κB nuclear translocation.22 As expected, cytokine-induced NF-κB translocation was abolished by Parkin transfection (Figure 1g and h). In contrast, si-Parkin promoted the translocation of NF-κB in response to cytokines (Supplementary Material, Supplementary Figure S2A and B). To confirm the role of Parkin on cytokine signaling, we checked the NF-κB transcription activity and JNK activation in Parkin null cell line, DLD-1.23 In this cell line, basal NF-κB transcription activity was elevated (Supplementary Material, Supplementary Figure S2C). However, Parkin transfection could block the basal activity as well as induction by cytokines (Supplementary Material, Supplementary Figure S2C). Similarly, p-JNK was elevated in DLD-1, comparing with HCT116 (Supplementary Material, Supplementary Figure S2D). Next, we tested the effect of sh-parkin on JNK activation and found that Parkin knockdown (Supplementary Material, Supplementary Figure S2E) could enhance the cytokine-inuduced JNK activation (Supplementary Material, Supplementary Figure S2F). To determine whether Parkin selectively suppresses cytokine signaling, we assayed the expression of p-JNK in response to oxidative stress and UV exposure. In contrast to the results observed with cytokine signaling, Parkin did not suppress p-JNK induced by oxidative stress or DNA damage (Supplementary Material, Supplementary Figure S3A). However, Parkin strongly suppressed the transcriptional activity of NF-κB in unstressed conditions (Supplementary Material, Supplementary Figure S3B). As reduction of κB-luc was obvious in H2O2 and UV-treated cells, we checked the viability and found that it would be resulted from reduction of cell viability (Supplementary Material, Supplementary Figure S3C). It is well known that JNK activation can be induced by ROS,24 we tested the involvement of ROS in Parkin-mediated JNK activation. However, treatment of N-acetyl L-cysteine (NAC), ROS scavenger, did not block the increase of p-JNK by si-Parkin (Supplementary Material, Supplementary Figure S3D). In addition, NAC did not alter the κB-luc activity (Supplementary Material, Supplementary Figure S3E and F). These results suggest that Parkin is not involved in ROS-mediated JNK activation.
Parkin regulates phospho-JNK (p-JNK). (a) Parkin suppresses JNK activation. The 293 cells that were transfected with either EV (empty vector) or Parkin were stimulated with 5 ng/ml of TNF-α or IL-1β for 30 min. Activated JNK (p-JNK) was suppressed by Parkin transfection. L.E and S.E indicates long exposure and short exposure, respectively. (b) Parkin suppresses JNK activation in the human neuronal cell line SH-SY5Y. Cells were transfected with Parkin for 24 h and incubated with IL-1β or TNF-α for 10 min. p-JNK and t-JNK indicate phosph-JNK and total JNK, respectively. (c) Reduction of IκB in Parkin-deficient lymphocytes. Because this cell line consists of immortalized Parkinson’s disease (PD) lymphocytes, Jurkat was used as the control cell line. (d) Parkin suppresses the transcriptional activity of NF-κB. NF-κB transcriptional activity was measured with an NF-κB luc vector, which was transfected into 293 cells. Promoter activity was determined with a luminometer after incubation with 5 ng/ml of TNF-α or IL-1β for 6 h. (e) Parkin regulates JNK activity. The 293 cell line was transfected with Parkin or si-Parkin for 24 h and triggered by TNF-α, IL-1β, and NGF-1 for 30 min. Parkin elimination can strongly induce the activation of JNK. (f) The effect of Parkin on NF-κB transcriptional activity in neuronal cells. SH-SY5Y cells were transfected with the indicated vectors and NF-κB luc for 24 h and incubated with the cytokines indicated for an additional 6 h. Statistical analysis was performed by Student’s t-test. P is probability. (g) Parkin blocks the translocation of NF-κB p65 into the nucleus. Cells were incubated with TNF-α for 6 h and divided into two fractions using a sub-cell fraction kit. Actin were used as cytoplasmic markers. (h) Immunofluorescence staining of NF-κB. Nuclear localization of NF-κB (green) induced by IL-1β was blocked by Parkin (red). DAPI (Blue) indicates the nucleus. White arrows indicate cytoplasmic NF-κB in Parkin-transfected cells.
Parkin Suppresses TRAF2/6 Expression
As TRAF2 and TRAF6 are essential mediators of cytokine signaling and can regulate JNK and NF-κB signaling,25 we assumed that Parkin would be related with TRAF2/6 pathway. In addition, deregulation of TRAF2-NF-κB signaling can promote tumor formation.26 To test our hypothesis, we measured the expression of TRAF2 and TRAF6 in Parkin- or si-Parkin-transfected cells, and we observed that the levels of both TRAF proteins were reduced by Parkin and slightly increased by si-Parkin in 293 and SH-SY5Y cells (Figure 2a and b). To confirm this result, we assayed the effect of Parkin on exogenous TRAF6, revealing that TRAF6 expression was clearly suppressed by Parkin in several types of cell lines (Figure 2c; Supplementary Material, Supplementary Figures S4A and B). However, transcripts of TRAF2/6 were not altered by Parkin and its mutants Supplementary Material, (Supplementary Figure S4C) as well as si-Parkin (Supplementary Material, Supplementary Figure S4D), indicating that Parkin regulated TRAF2/6 at post-translation level. Because Parkin is an E3 ligase, we measured the half-life of TRAF2 and TRAF6 in Parkin-transfected cells and found that expression of both proteins was diminished by Parkin (Figure 2d). Knockdown of Parkin extended the half-lives of TRAF2 and TRAF6 proteins (Figure 2d and e). Moreover, Parkin promoted the ubiquitination of TRAF6 and TRAF2 (Figure 2f; Supplementary Material, Supplementary Figure S4E). Although Parkin was able to associate with TRAF6 and AIMP2/p38, ubiquitination occurred more in TRAF6 than in AIMP2 (Figure 2f).12 In addition, TRAF6-induced NF-κB transcriptional activity was completely blocked by Parkin transfection (Figure 2g). In contrast, si-Parkin-induced NF-κB activity was blocked by the knockdown of TRAF6 (Figure 2h).
Parkin suppresses the expression of TRAF2 and TRAF6. (a) Parkin regulates TRAF2 and TRAF6 expression. The 293 cell line was transfected with Parkin or si-Parkin for 24 h, and the expression levels of TRAF2 and TRAF6 were measured. (b) In neuronal SH-SY5Y cells, Parkin can regulate TRAF2 expression. However, endogenous TRAF6 was not detected in this cell line. (c) Parkin also suppresses the exogenously supplied TRAF6 in N2A cells. Moreover, the expression levels of TRAF6 and Parkin were obviously reduced when they were co-transfected. (d) Parkin regulates the half-life of TRAF2 and TRAF6. Overexpression of Parkin strongly suppressed TRAF2 and TRAF6 expression, whereas si-Parkin extended the half-life of the proteins. The 293 cell line was transfected with Parkin or si-Parkin for 24 h and incubated with CHX for the indicated time to prevent de novo protein synthesis. (e) Quantitation of TRAF2 and TRAF6 expression. Protein half-life was determined by densitometry. Extension of TRAF2 and TRAF6 half-life by si-Parkin was obviously detected. (f) Parkin promotes ubiquitination of TRAF6 more strongly than of AIMP2. The 293 cell line was transfected with HA-Ub with Parkin or EV (empty vector) under ALLN-treated conditions. After IP with TRAF6 or AIMP2, Ub-conjugated TRAF6 or AIMP2 was detected by WB analysis with anti-HA antibodies. Interactions between Parkin and TRAF6 as well as Parkin and AIMP2 could be detected by WB. ‘Input’ indicates whole cell lysate in this analysis. Ubiquitination of TRAF2 can be observed in Supplementary Figure S4E. (g) Parkin blocks TRAF6-induced NF-κB activation. TRAF6-induced NF-κB activation was completely blocked by Parkin overexpression. The 293 cell line was co-transfected with the indicated vectors and NF-κB-luc vectors for 24 h. Statistical analysis was performed by Student’s t-test. P is probability. (h) Addition of si-TRAF6 blocks the NF-κB activation induced by si-Parkin. The elevated NF-κB activity normally induced by si-Parkin was blocked by si-TRAF6. Statistical analysis was performed by Student’s t-test. P is probability.
Direct Interaction of Parkin and TRAF2/6
Because TRAF2/6 seem to be targets of the Parkin E3 ligase, we checked the interactions between Parkin and TRAF2/6 using IP. Our results indicated that both proteins could interact with Parkin (Figure 3a and Supplementary Figure S4F). To identify the binding domain of Parkin, we performed a GST pulldown assay using Parkin fragments and found that the 141–237 region is responsible for the association with TRAF6 (Figure 3b). Because this region contains an IBR domain,12 we speculated that the IBR domain might be responsible for binding with TRAF2/6. Although the region from 385–416 was not covered by these fragments, this region also does not contain the functional domain. Thus, binding through the IBR domain seems to be plausible. To confirm this binding, we performed the GST pull down again, this time using recombinant TRAF6. As expected, the recombinant TRAF6 could also associate with Parkin (Supplementary Material, Supplementary Figure S4G). Using recombinant TRAF6 fragments, we determined that the N-terminal domain (RING domain) was responsible for binding to Parkin (Figure 3c). Next, we performed a ubiquitination assay to determine which domain is modified by Parkin. With this analysis, we confirmed that the N-terminal region of TRAF6 was modified by Parkin (Figure 3d). To confirm the direct interaction between Parkin and TRAF6, we performed the GST-pull down again using recombinant proteins and obtained the same result as when N-terminal TRAF6 was directly interacted with Parkin 141–416 region (Supplementary Material, Supplementary Figure S4H). To determine which specific residue on TRAF6 is ubiquitinated by Parkin, we generated 3-point mutant proteins. Because it has been reported that lysyl residues in the TRAF6 RING domain can be ubiquitinated,27 we replaced the lysyl residues in the RING domain with arginine (K91R, K96R, and K104R; Figure 3e). Although these residues can be auto-ubiquitinated,27 Parkin did not ubiquitinate two of these mutants (K91R and K96R; Figure 3e). To confirm the importance of ubiquitination of the K91 residue on the half-life of the TRAF6 protein, we delivered the recombinant proteins into EV- or Parkin-transfected cells and measured the protein stability. Compared with WT-TRAF6, K91R-TRAF6 showed resistance to Parkin-induced TRAF6 suppression (Figure 3f). In addition, addition of a proteasome inhibitor blocked the reduction of WT-TRAF6 in Parkin-transfected cells (Figure 3f), indicating that ubiquitination of the K91 residue (and perhaps also the K96 residue) is critical for Parkin-induced TRAF6 degradation.
Direct interactions between Parkin and TRAF2/6. (a) Interaction between TRAF2 and Parkin. IP analysis was performed to determine the binding between TRAF2 and Parkin. PUMA was used as a negative control. TRAF2 selectively interacted with Parkin. (b) The middle region of Parkin is responsible for its interaction with TRAF6. GST-Parkin deletion fragments were incubated with TRAF6-transfected 293 lysates for 30 min at room temperature. TRAF6 that associated with GST-tagged proteins was detected by WB analysis. Arrowheads indicate the expected sizes of the GST-fused Parkin fragments. (c) The N-terminal RING domain of TRAF6 is responsible for Parkin binding. Two types of recombinant TRAF6 (RING domain; 50–159 amino acids (aa), TRAF domain; 346-504 aa) were used for binding with Parkin, and the RING domain was co-immunoprecipitated with Parkin using a GST-pull down. (d) The N-terminal region of TRAF6 is ubiquitinated by Parkin. Recombinant TRAF6 proteins (N-terminal domain, TRAF6-N; C-terminal TRAF6, TRAF6-C) were incubated with EV- or Parkin-transfected 293 cell lysates (which had already been transfected with HA-Ub) for 6 h at room temperature. Ub-TRAF6 was detected by WB analysis with an HA antibody. (e) The TRAF6 residues R91 and R96 are ubiquitination sites. TRAF6 mutants (K91R, K96R and K104R) were incubated with 293 cell lysates that were co-transfected with HA-Ub and Parkin or EV. Ubiquitinated TRAF6 was detected by WB analysis with an HA antibody. The lysine residues (red) were replaced by arginine (lower panel). (f) TRAF6-K91R is resistant to Parkin-induced TRAF6 suppression. Recombinant TRAF6 proteins (WT and K91R) were delivered by liposomes into 293 cells that had been transfected with EV or Parkin. After 6 h, existing proteins were detected by WB analysis with an anti-GST antibody. When cells were treated with ALLN, GST-TRAF6-WT levels were not reduced in Parkin-transfected cells.
Parkin Mutants do not Degrade TRAF2/6
To address the involvement of Parkin’s E3 ligase activity on TRAF2/6 regulation, we next monitored the effect of mutant forms of Parkin (R42P and T240R) on TRAF2/6 expression. These mutants did not suppress the expression of TRAF2/6 (Figure 4a). In addition, mutant Parkin did not shorten the half-life of TRAF2/6 (Figure 4b). We also measured the NF-κB Luc-activity and found that Parkin mutants did not suppress NF-κB activity (Figure 4c). Next, we examined the interaction between the Parkin mutants and TRAF2/6 and monitored the ubiquitination of TRAF2/6 by mutant Parkin. To determine whether mutant forms of Parkin could interact with TRAF2/6, we re-performed the IP assay. Although these mutants possess the ability to bind with TRAF2/6 Supplementary Material, (Supplementary Figure S5A and B), they did not promote the ubiquitination of TRAF2 and TRAF6 (Figure 4d and e). This result is consistent with our previous finding that mutant Parkin did not reduce TRAF2/6 expression (Figure 4a and b). To confirm this, we investigated the exogenous protein stability. Exogenously delivered GST-TRAF6 was able to be retained in mutant Parkin-transfected cells (Figure 4f and Supplementary Material, Supplementary Figure S5C). In addition, mutant Parkin did not block the TNF-α induced NF-κB activation (Figure 4g). These results strongly suggest that TRAF2/6 are targets of the Parkin E3 ligase and that elevated expression or enforced TRAF2/6 signaling could be a cause of PD.
Mutant versions of Parkin do not suppress TRAF2/6 expression. (a) Two Parkin mutants (R42P and T240R) do not suppress TRAF2 and TRAF6. HCT116 cells were transfected with the vector indicated for 24 h. (b) Mutant Parkin shows an impairment of TRAF2 and TRAF6 degradation. Compared with wild-type (WT)-Parkin, R42P-Parkin did not promote the turnover of TRAF2 and TRAF6. (c) Mutant Parkin cannot suppress NF-κB activity. Neither mutant Parkin suppressed the transcriptional activity of NF-κB. HCT116 cell lines were transfected with wild-type or mutant Parkin vectors for 24 h. Statistical analysis was performed by Student’s t-test. P is probability. (d) Only WT-Parkin promotes TRAF6 ubiquitination. However, R42P-Parkin was able to interact with TRAF6. The 293 cells were transfected with WT or R42P-Parkin and incubated with ALLN (20 μM) for 4 h. The cell lysates were incubated with a TRAF6 antibody, and precipitated proteins were analyzed by WB. (e) Mutant Parkin does not cause TRAF2 ubiquitination. Under the same conditions, IP analysis was performed with a TRAF2 antibody. (f) Mutant Parkin does not reduce exogenously supplied TRAF6 protein. After transfection with WT or mutant Parkin for 24 h in HCT116, GST-TRAF6 protein was delivered using liposomes. After 6 h, the amount of protein was determined by WB analysis with anti-GST antibody. Time-dependent reduction of TRAF6 by WT-parkin could be observed in Supplementary Figure S5C. (g) Mutant forms of Parkin do not suppress the induction of NF-κB activation in response to TNF-α. The 293 cells were used for a luciferase assay. Statistical analysis was performed by Student’s t-test. P is probability.
Parkin Protects Against Cytokine-Induced Cell Death
Because PD is evoked by cell death in the SN,1, 2 we examined the effect of Parkin on cell death. Overexpression of Parkin suppressed TNF-α-induced cell death (Figure 5a), whereas si-Parkin reduced cell viability (Figure 5b). Moreover, elimination of TRAF2 or TRAF6 blocked the si-Parkin-induced cell death (Figure 5c; Supplementary Material, Supplementary Figure S6A). In neuronal cells, we obtained a similar result in that si-Parkin-mediated cell death was rescued by si-TRAF6 (Figure 5d). To confirm this finding and to determine whether the reduction of cell viability was achieved by apoptosis, we monitored the release of cytochrome c in SH-SY5Y cells. The si-Parkin-induced cytochrome c release was completely blocked by si-TRAF6 (Figure 5e and f). We also observed a similar effect of si-TRAF6 on IL-1β-induced apoptosis (Figure 5f and Supplementary Material, Supplementary Figure S6B).
Parkin blocks cytokine-induced cell death. (a) Parkin protects against TNF-α induced cell death in 293 cells. After a 24 h transfection with Parkin or EV, cells were incubated with CHX and TNF-α for 12 h. TNF-α-induced cell death was completely blocked by Parkin transfection. An MTT assay was used to determine the cell viability. Statistical analysis was performed by Student’s t-test. P is probability. (b) Decreased cell viability from si-Parkin. The 293 cells were transfected with si-Parkin or si-control for 24 h and incubated with CHX (100 μg/ml) for 3 h. After treatment with TNF-α or IL-1β for 12 h, cell viability was determined with an MTT assay. Statistical analysis was performed by Student’s t-test. P is probability. (c) Use of si-TRAF6 blocks the Parkin-deficiency-induced apoptosis. TNF-α or IL-1β-induced cell death in si-Parkin-transfected cells was blocked by si-TRAF2 co-transfection. HCT116 cells were transfected with the indicated si-RNA for 24 h and incubated with TNF-α or IL-1β for 12 h in the presence of CHX. Cell viability was determined with an MTT assay. Statistical analysis was performed by Student’s t-test. P is probability. (d) Use of si-TRAF6 also suppresses the si-Parkin-induced cell death in neuronal SH-SY5Y cells. MTT was performed under the conditions described above. Statistical analysis was performed by Student’s t-test. P is probability. (e) Cytochrome c (Cyto. C) release in TNF-α-treated SH-SY5Y cells. Cells were transfected with the indicated si-RNA for 24 h, were incubated with TNF-α for 6 h with CHX, and were then stained with a cytochrome c antibody. (f) Use of si-TRAF6 blocks the apoptosis otherwise induced by si-Parkin. Cells stained with diffused cytochrome c in the cytosol and nucleus were examined, and the apoptotic cells were counted. At least 200 cells per sample were counted in three independent experiments.
Increase of TRAF6 in MPTP-Induced PD Mouse Model
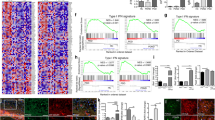
To further examine the physiological relevance between TRAF6 and PD, we generated a mouse PD model using MPTP-injected mice.28 It has been confirmed that MPTP induces the loss of tyrosine hydroxylase-positive neurons in the SN (Figure 6a and Supplementary Figure S7A). In addition, the rotarod performance test showed that MPTP injection led to the impairment of movement ability (Supplementary Material, Supplementary Figure S7B). In this experiment, we were able to observe the induction of TRAF6 in MPTP-treated neurons from two independent experiments (Figure 6b and c). However, GFAP-positive glial cells did not show the induction of TRAF6 in response to MPTP (Figure 6b and c). Thus, we measured the expression of TRAF6 and IκB from 24-h-MPTP-injected mouse SN lysates. Consistent with our hypothesis and previous results (Figure 6b and c), increased TRAF6 and reduced levels of IκB were detected in the MPTP-injected mice (Figure 6d). This result suggests that the induction of TRAF6 may be essential for dopaminergic neuronal cell death.
Elevated expression of TRAF6 in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced mouse Parkinson’s disease (PD) model. MPTP-induced tyrosine hydroxylase (TH)-positive neuron loss. MPTP-treated mouse brain slices were stained with a TH antibody. Arrows indicate reduction of TH-positive neurons. (b) Increased TRAF6 expression in MPTP-treated SN neurons. In the same tissues, staining for TRAF6 (red) was performed with co-staining with GFAP (green; marker of glial cells). TRAF6 expression was increased in neurons. (c) The same result from a different mouse tissue. (d) Increased TRAF6 protein levels in PD mouse tissues. To confirm the induction of TRAF6, we extracted the protein from the SN of a mouse brain and examined the levels of TRAF6 expression. Consistent with the results observed in IF, TRAF6 expression was elevated in the MPTP-treated samples. In addition, we observed the reduction of IκB.
Elevated Expression of TRAF6 in Human PD Tissues
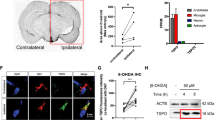
To address the pathological relevance of TRAF6 and Parkin, we obtained 15 human PD tissues (SN region) from the Victorian Brain Bank Network and analyzed the expression of TRAF6 and Parkin through IHC. In normal tissues, TRAF6 expression was weakly or negatively detected (Figure 7a and Supplementary Figure S8). In contrast, nine cases showed elevated expression levels of TRAF6 (Figure 7a and Supplementary Material, Supplementary Figure S9). Moreover, TRAF6 expression showed a reverse relationship with Parkin (Figure 7a; Supplementary Material and Supplementary Figure S8 and S9). However, some tissues showed an aggregation of TRAF6 with Parkin-positive staining (Supplementary Material, Supplementary Figure S9; Figure 7b (right panel; Code # 437 and # 566)). We speculate that these instances could result from Parkin mutation, as mutant Parkin can interact with TRAF2/6 but cannot eliminate it. To confirm this speculation, we examined the expression pattern of TRAF6 by Parkin transfection and found that Parkin mutants could induce TRAF6 aggregation in the cytosol as observed in the IHC of human tissues (Figure 7b). Although we could not perform a mutation analysis using these samples, it is plausible that these tissues may contain Parkin mutations.
Reciprocal expression of Parkin and TRAF6 in Parkinson’s disease (PD) tissues. (a) IHC staining in human PD and normal tissues. In normal tissues, Parkin expression was detected (brown). In addition, several PD tissues also stained for Parkin (left panels). In these samples, TRAF6 expression was not detected. In contrast, other tissues showed elevated expression of TRAF6 and reduction of Parkin (right panels). Black granules in neurons are the dopaminergic granules (arrows). PD tissues are well known for showing the loss of dopaminergic granules. (b) Parkin mutants induce TRAF6 aggregation. To test our hypothesis that Parkin mutants induce TRAF6 aggregation in the cytosol and explain previous results (right panel; PD #437 and #566), we stained for TRAF6 in Parkin-mutant-transfected cells and found that both Parkin mutants (R42P and T240R) evoke TRAF6 aggregation in the cytosol (green). However, Parkin aggregation was detected only in the R42P mutant.
DISCUSSION
Because PD is induced by neural apoptosis, Parkin, which is deleted in juvenile types of Parkinson disease,2 seems to block this cell death and is therefore called an anti-apoptotic gene. However, deletion of Parkin has also been reported in human cancer tissues.13, 14 These findings are inconsistent with our first premise. To resolve these conflicting data, we proposed the hypothesis that Parkin would be linked to inflammation, which can promote cell death and can also induce cancer. Indeed, chronic inflammation in the colon or epithelial tissues can promote the formation of cancer. In contrast, inflammation in the brain or the CNS can induce tissue damage and neuronal cell death.21
In this study, we showed that Parkin can suppress cytokine-induced JNK activation and NF-κB signaling (Figure 1). Considering the fact that loss of CYLD induces tumors by deregulation of TRAF2-NF-κB signaling,26 loss of Parkin may also promote tumor formation. In addition, we showed that TRAF2 and 6 are direct targets of Parkin. Parkin can interact with TRAF2/6 and promote their ubiquitination (Figure 2). Although many types of proteins are proposed as targets or binding partners of Parkin,2 they do not fully explain the phenotype of Parkin-related PD and the relationship with cancer. However, TRAF2/6 are linked to inflammation, which is critical for cell death and cancer. Thus, Parkin-mediated TRAF2/6 suppression may explain Parkin’s relationship with PD and cancer.
We also provide evidence for the relevance of TRAF2/6 in the loss of Parkin-induced cell death. Elimination of TRAF2/6 can block the cytokine-induced neuronal cell apoptosis, which is enhanced by Parkin knockdown (Figure 4). Indeed, we found elevated expression of TRAF2/6 in human PD tissues (Figure 7) and in our MPTP-induced mouse PD model (Figure 6). In addition, we revealed that mutant Parkin did not promote the degradation of TRAF2/6 but did induce TRAF6 aggregation in the cytosol (Figure 7b). These results strongly suggest that TRAF2/6 is a physiological target of the Parkin E3 ligase.
Our results show that the E3 ligase Parkin can suppress TRAF2/6 signaling, which induces chronic inflammation through NF-κB activation and apoptosis via the JNK/stress signaling pathway (Figure 8). This function explains frequent Parkin mutations found in human cancers.13, 14 Because cancer cells originate from replaceable epithelial cells, chronic inflammation can trigger cell proliferation, which will contribute to the progression of the cancer. Thus, Parkin-deficient cells will be more sensitive to inflammation. In contrast, neuronal cells cannot be regenerated by stem cells. Thus, chronic inflammation may induce continuous cell loss, which will induce neurodegenerative diseases such as PD. The prevention of inflammation or reduction of TRAF2/6-mediated signaling in neuronal cells could be a putative therapeutic strategy.
Model of putative pathways. Under physiological conditions, Parkin can suppress excessive activation of TRAF2/6 through poly-ubiquitination and proteasomal degradation. When Parkin is deleted or mutated, activated TRAF2/6 promotes JNK activation, which may lead to cell death in neurons, and NF-κB activation, which will enhance cytokine expression (positive regulation). Under the same genetic conditions, epithelial cells of the colon can progress into a tumor because of NF-κB signaling.
References
Thomas B, Beal MF . Parkinson’s disease. Hum Mol Genet 2007;16:183–194.
Dawson TM, Dawson VL . The role of parkin in familial, and sporadic Parkinson’s disease. Mov Disord 2010;25:S32–S39.
Tansey MG, Goldberg MS . Neuroinflammation in Parkinson’s disease: its role in neuronal death, and implications for therapeutic intervention. Neurobiol Dis 2010;37:510–518.
McGeer EG, Klegeris A, McGeer PL . Inflammation, the complement system, and the diseases of aging. Neurobiol Aging 2005;26:S94–S97.
Block ML, Hong JS . Microglia, and inflammation-mediated neurodegeneration: multiple triggers with a common mechanism. Prog Neurobiol 2005;76:77–98.
Farrer M, Chan P, Chen R et al. Lewy bodies, and parkinsonism in families with parkin mutations. Ann Neurol 2001;50:293–300.
Sasaki S, Shirata A, Yamane K et al. Parkin-positive autosomal recessive juvenile Parkinsonism with alpha-synuclein-positive inclusions. Neurology 2004;63:678–684.
Ishikawa A, Takahashi H . Clinical, and neuropathological aspects of autosomal recessive juvenile parkinsonism. J Neurol 1998;245:P4–P9 (suppl 11).
Shimura H, Hattori N, Kubo S et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 2000;25:302–305.
Dong Z, Ferger B, Paterna JC et al. Dopamine-dependent neurodegeneration in rats induced by viral vector-mediated overexpression of the parkin target protein, CDCrel-1. Proc Natl Acad Sci USA 2003;100:12438–12443.
Imai Y, Soda M, Inoue H et al. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 2001;105:891–902.
Ko HS, von Coelln R, Sriram SR et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci 2005;25:7968–7978.
Poulogiannis G, McIntyre RE, Dimitriadi M et al. PARK2 deletions occur frequently in sporadic colorectal cancer, and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA 2010;107:15145–15150.
Veeriah S, Taylor BS, Meng S et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma, and other human malignancies. Nat Genet 2010;42:77–82.
Frank-Cannon TC, Tran T, Ruhn KA et al. Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J Neurosci 2008;28:10825–10834.
Villarán RF, Espinosa-Oliva AM, Sarmiento M et al. Ulcerative colitis exacerbates lipopolysaccharide-induced damage to the nigral dopaminergic system: potential risk factor in Parkinson’s disease. J Neurochem 2010;114:1687–1700.
Yang DJ, Chung JY, Lee SJ et al. Slug, mammalian homologue gene of Drosophila escargot, promotes neuronal-differentiation through suppression of HEB/daughtherless. Cell Cycle 2010;9:2789–2802.
Lee SH, Shen GN, Jung YS et al. Antitumor effect of novel small chemical inhibitors of Snail-p53 binding in K-Ras-mutated cancer cells. Oncogene 2010;29:4576–4587.
Hunot S, Dugas N, Faucheux B et al. FcepsilonRII/CD23 is expressed in Parkinson’s disease, and induces, in vitro, production of nitric oxide, and tumor necrosis factor-alpha in glial cells. J Neurosci 1999;19:3440–3447.
Orr CF, Rowe DB, Mizuno Y et al. A possible role for humoral immunity in the pathogenesis of Parkinson’s disease. Brain 2005;128:2665–2674.
Hirsch EC, Hunot S . Neuroinflammation in Parkinson’s disease: a target for neuroprotection? Lancet Neurol 2009;8:382–392.
Wajant H, Pfizenmaier K, Scheurich P . Tumor necrosis factor signaling. Cell Death Differ 2003;10:45–65.
Ikeuchi K, Marusawa H, Fujiwara M et al. Attenuation of proteolysis-mediated cyclin E regulation by alternatively spliced Parkin in human colorectal cancers. Int J Cancer 2009;125:2029–2035.
Mansat-de MV, Bezombes C, Quillet-Mary A et al. Implication of radical oxygen species in ceramide generation, c-Jun N-terminal kinase activation and apoptosis induced by daunorubicin. Mol Pharmacol 1999;56:867–874.
Li H, Lin X . Positive, and negative signaling components involved in TNF-alpha-induced NF-kappaB activation. Cytokine 2008;41:1–8.
Kovalenko A, Chable-Bessia C, Cantarella G et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 2003;14:801–805.
Lamothe B, Besses A, Campos AD et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IkB kinase activation. J Biol Chem 2007;282:4102–4112.
Pan J, Qian J, Zhang Y et al. Small peptide inhibitor of JNKs protects against MPTP-induced nigral dopaminergic injury via inhibiting the JNK-signaling pathway. Lab Invest 2010;90:156–167.
Acknowledgements
This research was supported by a grant from the National Research Foundation of Korea to NC Ha (2011-0015420) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0065237, BJ Park).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Parkin, an E3 ligase, suppresses inflammation and cytokine-induced cell death by promoting the proteasomal degradation of TNF-α receptor associated factor (TRAF) 2/6. Because TRAF6 is elevated in Parkin-deficient human Parkinson's disease samples, these results may provide the basic pathological mechanism of Parkinson's disease.
Supplementary information
Rights and permissions
About this article
Cite this article
Chung, JY., Park, H., Lee, SJ. et al. Elevated TRAF2/6 expression in Parkinson’s disease is caused by the loss of Parkin E3 ligase activity. Lab Invest 93, 663–676 (2013). https://doi.org/10.1038/labinvest.2013.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2013.60
Keywords
This article is cited by
-
Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities
Cell Death & Differentiation (2021)
-
Structural feature of TRAFs, their related human diseases and therapeutic intervention
Archives of Pharmacal Research (2021)
-
The impact of proteostasis dysfunction secondary to environmental and genetic causes on neurodegenerative diseases progression and potential therapeutic intervention
Environmental Science and Pollution Research (2020)
-
Loss of Microglial Parkin Inhibits Necroptosis and Contributes to Neuroinflammation
Molecular Neurobiology (2019)
-
TRAF Molecules in Inflammation and Inflammatory Diseases
Current Pharmacology Reports (2018)