Abstract
The diagnosis of Marfan syndrome (MFS) remains challenging despite the 2010 revision to Ghent nosology criteria, and there is a lack of published information regarding FBN1 genotype associations in patients since the update in Ghent criteria. Applying revised Ghent criteria, we reviewed consecutive proband cases (n=292) submitted for FBN1 sequencing. Testing yielded 207 pathogenic or likely pathogenic FBN1 variants, with 114/207 (55%) missense, 67/207 (32%) non-sense or frameshift, and 28/207 (13%) splicing. There were 130 novel FBN1 variants predicted as pathogenic or likely pathogenic (n=109) or variant of undetermined significance (n=21). Of the 104 patients who met 2010 revised Ghent criteria, 87/104 (82%) had a pathogenic or likely pathogenic variant. There was a significantly lower frequency of missense variants (41 vs 89%; P<0.0001) observed in the Ghent-positive (vs Ghent-negative) patients, and this association held true in age-based groupings. Previously described genotype associations with ectopia lentis and early onset/‘neonatal’ MFS were confirmed in our cohort. Overall, our study points to the imperfect nature of relying solely on clinical criteria to diagnose MFS as well as the potential importance of truncating/splicing variants in Ghent-positive cases. Furthermore, the description of numerous novel variants and associated clinical findings may be useful for future clinical interpretation of FBN1 genotype in patients with suspected MFS.
Similar content being viewed by others
Introduction
Marfan syndrome (MFS), due to pathogenic variants in FBN1, is an autosomal dominant connective tissue disorder typically involving the ocular, skeletal and cardiovascular systems; but may less frequently can involve the skin, integument, lung, muscle and adipose tissue.1 Mutations in FBN1 are numerous and have been found across the gene. Many FBN1 pathogenic variants are unique to individual families. Missense variants are the most common type of FBN1 variant, with the majority of these being cysteine substitutions.2 Approximately 25% of FBN1 pathogenic variants are de novo variants.3 There are very few strong genotype–phenotype correlations, with the exception of the association of early onset, rapidly progressive (previously termed ‘neonatal’) MFS and FBN1 variants in exons 24 through 32. A higher probability of ectopia lentis (EL) in patients with missense variants substituting or creating a cysteine residue and a lower probability of EL in patients with a protein truncating variant has also been observed.2, 4, 5, 6, 7
Genetic analysis of FBN1 is important to confirm a suspected diagnosis of MFS or related fibrillinopathy, as well as for potential elucidation of genotype–phenotype correlations. In the most recent diagnostic criteria published in 2010 (the revised Ghent Nosology criteria for MFS),8 a diagnosis of MFS can be made when a causal FBN1 variant is identified in the presence of aortic dissection or an aortic root Z-score of ⩾2. Alternatively, a causal FBN1 variant in the setting of EL and known aortic root dilation also confirms the diagnosis. A diagnosis can be made in the absence of a causal FBN1 variant as well, taking into account factors such as systemic score, aortic root dilation z-score, EL and family history. The type of FBN1 variant identified and its likelihood of being pathogenic are recognized as important factors when making a diagnosis of MFS, with de novo (in the absence of family history), non-sense, frameshift, splicing and missense substitutions of conserved residues considered more likely to be pathogenic than other missense variants.
An improved or relatively unchanged diagnostic yield for MFS has been demonstrated in light of the 2010 revised Ghent criteria.9, 10 However, there is a scarcity of published information regarding genotype associations in patients who fulfill revised Ghent criteria. Furthermore, the enhanced genomic knowledge in recent years has allowed for improved classification of genetic variants with regards to pathogenicity. On the basis of these recent changes, we sought to investigate genotype and phenotype relationships in a cohort of consecutive probands who underwent FBN1 analysis due to suspected or clinically confirmed MFS. The purpose of this study was three-fold: (1) to assess the relationship of FBN1 analysis to MFS diagnosis in light of the revised Ghent criteria; (2) to explore and report FBN1 genotype–phenotype relationships in our cohort; and (3) to describe novel FBN1 variants associated with the MFS phenotype.
Materials and methods
Patient samples and clinical information
Proband samples (n=292) with a suspected or confirmed diagnosis (utilizing 2010 revised Ghent criteria) of MFS were included in this study. Proband samples were received by our clinical reference laboratory (Mayo Medical Laboratories, Rochester, MN, USA) from local, national and international sources. Each patient was examined by his or her referring physician. For Mayo Clinic patients, phenotypic information was extracted from the patients’ electronic medical record. For patients external to Mayo Clinic, phenotypic information was provided by the referring provider via a requisition form specific to MFS and related disorders that included age, gender, suspected diagnosis, family history and phenotypic features, including those related to the 2010 revised Ghent nosology criteria. Ghent-positive or -negative status was applied utilizing clinical and genotypic information. The study was approved by the Mayo Foundation Institutional Review Board.
FBN1 sequencing
Genomic DNA was extracted from EDTA-anticoagulated whole blood. All 65 exons of FBN1 and a minimum of 20 basepairs (bp) of intronic DNA flanking each exon were amplified by PCR. Amplification was performed using a common master mix containing Platinum Taq DNA Polymerase, 10 × PCRx Enhancer System, 10 × PCR Buffer (-MgCl), MgSO4 (all from Invitrogen, Carlsbad, CA, USA), and a 10 mM dNTP mixture (Roche, Indianapolis, IN, USA). Master mix, forward and reverse primers were combined with genomic DNA and amplified by 35 cycles of PCR (30 s at 95 °C; 30 s initially at 68 °C then decreased by 0.5 °C each cycle, with the last 20 cycles performed at 60 °C; and 1 min extension at 72 °C, with a final 10 min extension at 72 °C). Amplicons were bi-directionally sequenced using Big Dye Terminator technology on an ABI 3730 system (Applied Biosystems, Foster City, CA, USA). Sequence analysis was done using the Mutation Surveyor software (SoftGenetics, State College, PA, USA) and visual inspection.
Classification of sequence variants
FBN1 variant nomenclature was based on RefSeq NM_000249.3. Variants were classified based on a five-level classification system: pathogenic, likely pathogenic, variant of undetermined significance (VUS), likely benign or benign. FBN1 variants were analyzed for pathogenicity based on criteria that included (a) whether or not they were previously reported with disease and/or had associated functional studies, (b) nature of variant (for example, missense and non-sense), (c) location of variant (for example, critical cysteine residue of calcium binding epidermal growth factor-like (cbEGF-like) domain), (d) frequency of variant in the Exome Variant Server (EVS) database and dbSNP.
Variants were classified as pathogenic if they were non-sense point mutations, frameshift insertions/deletions, and variants involving the splice donor (intron +1G or +2T) or splice acceptor (intron −1G or −2A). Variants were classified as likely pathogenic if the variant affected or created a cysteine residue in a cbEGF-like or transforming growth factor-beta binding protein (TB) domain, if it affected a known consensus/critical residue (for example, the critical glycine at position 3 between cysteine 2 and cysteine 3 in the cb-EGF-like domain),11, 12, 13 if there was a previous literature report describing a negative impact of the variant on protein function, and/or if the variant was determined to be de novo or otherwise likely pathogenic based on family studies. Variants were classified as benign or likely benign if they had frequency >0.39% (or sometimes lower depending on other factors) in the EVS database and dbSNP, had functional studies demonstrating no deleterious effect, were not at highly conserved residues or located in a highly variable region without a known function, were deeply intronic (> +/−20 bp from exon-intron boundary) and not predicted to impact splicing, and/or were not supported to be pathogenic based on in silico analyses. Variants were classified as VUS if they could not be categorized as one of the above and/or due to a lack of sufficient functional studies or frequency information.
Variants not definitively known to be pathogenic underwent in silico splicing analyses utilizing Alamut version 2.2 (Interactive Biosoftware, Rouen, France) integrated software tools GeneSplicer, MaxEntScan, NNSPLICE, SpliceSiteFinder-like and Human Splicing Finder. Additional online tools accessed through Alamut (MutationTaster, Align GVGD, PolyPhen-2 and SIFT) were utilized to analyze missense variants. Alamut was also utilized to derive allele frequency information via the EVS, dbSNP, 1000 Genomes and other online publicly available resources. Searches were performed online and in databases (for example, Human Gene Mutation Database) to aid in determination of novelty and significance of findings. Family testing was performed in some cases to aid in determination of clinical significance of specific variants.
Statistical analyses
Statistical significance was calculated by GraphPad software (www.graphpad.com), utilizing Fisher’s exact test to calculate P-values.
Results
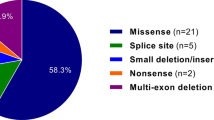
Testing yielded 280 FBN1 variants that were classified as pathogenic, likely pathogenic, VUS or likely benign (Table 1). Overall, 207 pathogenic and likely pathogenic variants were observed (Supplementary Table 1). The median age at testing for individuals with pathogenic or likely pathogenic variants was 17 years (average age at testing, 22 years). The frequency of the pathogenic and likely pathogenic variant type is depicted in Figure 1a and listed in Table 1.
(a) Frequency of pathogenic/likely pathogenic variant type; (b) specific type of missense variant in relation to domain and impact on critical/conserved FBN1 amino-acid residues. cbEGF-like, calcium binding EGF-like domain; Cys, cysteine residue; D-I/T/E/V/L-N/D-E, 4 amino-acid consensus region in 5′ cbEGF-like domain; TB, TGFβ binding protein domain.
Missense variants were the major type of pathogenic/likely pathogenic variant, and they were categorized based on potential functional impact (Figure 1b). The majority (75/114; 66%) of these 114 missense variants occurred in the cbEGF-like domains. For missense variants that occurred within the cbEGF-like domain, 91% of them impacted a known cbEGF-like highly conserved residue, such as altering one of the six critical cysteine (Cys) residues in the cbEGF-like domain. Specifically, in the cbEGF-like domain, 36% (31/87) variants altered one of the six critical cysteines, 21% (18/87) variants created a new Cys, 25% (22/87) altered one of the residues in the four amino-acid D-I/T/E/V/L-N/D-E consensus region, and 9% (8/87) affected another critical residue, such as a calcium binding residue or a critical glycine. In all, 11% of the missense variants (12/114) occurred in the TB domain and 58% of these (7/12) either created a cysteine or altered one of the 8 critical cysteines in a TB domain. Twenty-three percent of the missense variants (26/114) did not impact a known critical residue, but were considered as pathogenic or likely pathogenic due to previous reports that demonstrated functional impact and/or because of segregation with disease in the family.
Ghent criteria status
Taking into consideration phenotypic features and genotype, there were 104 patients who were determined to meet 2010 revised Ghent criteria. For genotype, pathogenic and likely pathogenic variants meeting criteria for causal variant as per the 2010 revised Ghent criteria were included in the categorization of Ghent status.8 Of the patients whom met Ghent criteria, 87 (82%) had a pathogenic or likely pathogenic variant (Table 1). The majority of these variants (51/87; 59%) were protein truncating (that is, non-sense or frameshift) or splicing. The other 36 variants (41%) were missense. Overall, there was a lower frequency of missense variants (41 vs 89%; P<0.0001) observed in the Ghent-positive patients compared with the Ghent-negative patients (Table 1; Figure 2). The frequency of missense variants affecting critical cysteine residues was not statistically significantly different in Ghent-positive patients as compared with Ghent-negative patients (64 vs 47%, P=0.1311).
Since some MFS features may not be apparent in younger aged patients, patients were separated into two groups based on age. Adults were considered to be ⩾20 years old, whereas young individuals were categorized as <20 years old.8 The pattern of variant type observed in adults vs young individuals with and without meeting Ghent criteria was very similar to what was observed in the Ghent-categorized cohort overall (Table 1; Figure 2). The highest frequency of missense variants was observed in patients <20 years old (whether Ghent criteria positive or negative). However, there were no statistically significant differences in missense variant frequency between age groups in the Ghent-positive or Ghent-negative patients.
There were 17 patients whom met Ghent criteria, but whom did not have an identifiable FBN1 causal variant by sequence analysis. Of this group of patients, the majority (11/17; 65%) had aortic root Z-score ⩾2 and systemic score ⩾7 points (but no family history). Additionally, 4/17 had aortic root Z-score ⩾2 and systemic score ⩾7 points (plus family history); 1/17 had aortic root Z-score ⩾2 and EL, and 1/17 had systemic score ⩾7 points and family history.
Novel pathogenic and likely pathogenic variants
Of the 207 pathogenic and likely pathogenic variants, 53% (109/207) were novel, previously unreported variants (Table 2). For the novel variants, 52% (57/109) were missense, 15% (16/109) were non-sense, 19% (21/109) were frameshift and 14% (15/109) were splicing variants. Within the missense variant sub-grouping, the majority (81%; 46/57) of these occurred in the cbEGF-like domains, usually at critical residues. Seventeen of the novel missense variants were tested in family members of the proband to help determine whether the variant segregated with disease and/or was not present in unaffected family members (Tables 3 and 4).
Novel splicing variants
As described above, 14 novel splicing variants were observed, and most of them occurred in the critical intronic donor and acceptor splicing regions (one and two bases upstream and downstream of the exon, respectively). Two novel splicing variants that were considered to be likely pathogenic were observed outside this region. Given that these two variants would questionably impact splicing, we performed familial and in silico analyses to help ascertain the pathogenicity of the variants, as detailed below. We have also included the clinical details of these two probands below in order to further provide support for the potential pathogenicity of these two variants.
The first novel splicing variant outside the critical splice junction region, in patient 29, a 2-year-old Asian (Indian) male, was observed to harbor the c.2114-3C>A variant. This patient had bilateral EL, bicuspid aortic valve with mild insufficiency, reduced upper to lower segment ratio, armspan to height ratio >1.05, pes planus, joint hypermobility and highly arched palate with crowded teeth. There was no family history of MFS and his unaffected parents both tested negative for the variant thus making this variant a likely de novo variant in the proband, assuming correct assignment of paternity. Four in silico splice prediction programs (SpliceSiteFinder-like, MaxENTScan, GeneSplicer and Human Splicing Finder) predict that this variant reduces splicing at the canonical splice acceptor site for intron 16.
The second splicing variant outside the critical splice junction region was c.4583-5A>G in a 22-year old Caucasian male (patient 64). This patient met Ghent criteria and had a positive wrist sign, pectus carinatum, hindfoot valgus deformity, pes planus, facial features, skin striae, possible myopia and a dilated aortic root. Similar to the previous patient described above, there was no family history of MFS and his unaffected parents both tested negative for the variant, thus making this variant a likely de novo occurrence in the proband, assuming correct assignment of paternity. Five in silico splice prediction programs (SpliceSiteFinder-like, MaxENTScan GeneSplicer, NNSPLICE and Human Splicing Finder) predict that this variant creates a new splice acceptor site in intron 36.
Novel VUSs
Thirty-three probands had VUSs identified (Table 3). Some of the VUSs were observed in multiple probands; therefore, there were 30 unique VUSs observed, of which 21 were novel, previously unreported variants. The majority of the VUSs (70%; 21/30) were missense variants with eight of the VUSs occurring in the intronic regions. There were four exonic missense VUSs that were predicted to potentially impact splicing: p.G899V (c.2696G>T), p.N1168S (c.3503A>G), p.G2506S (c.7516G>A) and p.G2618R (c.7852G>A).
Familial testing
Nineteen probands with novel missense variants and novel or previously reported VUSs had additional family testing performed to help elucidate the significance of the variant identified (Table 4). Seventeen of these variants were missense and two were potential splicing variants. As described above, the two potential splicing variants were both determined to be de novo, assuming correct assignment of paternity. Of the seventeen missense variants, family testing in nine of the cases supported the classification of likely pathogenic. There were eight inconclusive cases (p.C209R, p.M977R, p.N1168S, p.I1175V, p.G2116A, p.P2471R, p.D2569Y and p.L2778P) where either the variant did not track with symptomatic family members, or not enough information was provided to determine whether the variant tracked with disease, or too few family members were tested to substantiate pathogenicity or lack thereof.
Neonatal region
We tested nine patients with a severe form of MFS present from birth, previously termed neonatal MFS. The age range of patients tested was 9–525 days old (mean, 91 days old). As previously reported, most patients with this presentation have causal variants in exons 24–32, especially missense variants in exons 24–32 and variants resulting in exon skipping of exon 31 or 32.14, 15 Of the nine patients that we tested for variants in exons 24–32, four of these patients were observed to have a pathogenic variant. One of the variants occurred in exon 25 (p.I1048T), two of the variants occurred at cysteine residues in exon 26 (p.C1074S (novel) and p.C1086Y) and one variant occurred at the splice donor site of intron 32 (c.4087+1G>A). As is typical with early onset, rapidly progressive MFS, no family history of MFS was reported for any of these patients.
Ectopia lentis
EL has been reported as more likely associated with missense variants involving cysteine residues than with other types of missense, exon skipping and premature truncating variants.2, 4, 5, 6, 7, 16 It has also been reported that patients with EL have pathogenic variants clustering in the first 15 exons of FBN1,6, 17 although this was not observed in another study.4 In our cohort, there were 65 individuals with reported EL, and 50 of them tested positive for a pathogenic or likely pathogenic variant, with 39/50 (78%) being a missense variant (Table 1). Overall, 26/50 (52%) either altered or created a cysteine residue (vs 14% for cohort without reported EL; P<0.0001), 10/50 (20%) were truncating variants (vs 51% cohort without reported EL; P<0.0001), and 17/50 (34%) occurred in exons 1–15 (vs 20% cohort without reported EL; P=0.0027). To reduce the risk of bias due to incomplete reporting of phenotype in our cohort, we excluded mutation-positive patients whom did not have accompanying phenotypic information (n=27) from this analysis.
Discussion
With the revision to Ghent criteria in 2010, few studies have been performed to assess the association with FBN1 genetic analysis and MFS diagnosis in light of the revised criteria. Before the revised Ghent criteria, it had been observed that for individuals who met Ghent criteria, the FBN1 causal variant detection rate ranged from 66 to 93%.4, 5, 6, 18, 19 A more recent smaller study observed an 80% FBN1 causal variant detection rate in 24/30 patients who fulfilled 2010 revised Ghent criteria.20 Another study observed that genotype information changed the final diagnosis in more patients who met revised Ghent criteria as compared with original Ghent criteria, thus pointing to the importance of genotype analysis in suspected MFS cases.21
To help address the question of extent of correlation with FBN1 genotype and revised Ghent criteria, we reviewed 292 proband cases that underwent FBN1 sequence analysis. In our cohort, there were 104 patients who were determined to meet 2010 revised Ghent criteria, and 87 (82%) had a pathogenic or likely pathogenic FBN1 variant. Thus, our causal variant detection rate in Ghent-positive patients was similar to that reported in previous studies. Additionally, of the 207 patients with a pathogenic or likely pathogenic variant, 55 of them did not meet Ghent criteria. This speaks to the imperfect nature of relying solely on clinical criteria to diagnose MFS, and the importance of the genetic evaluation of patients with suspected MFS.
An FBN1 causal variant was not identified in 16% of Ghent-positive patients in our study. This may be due, at least in part, to the phenotypic overlap with other genetic disorders such as Loeys-Dietz syndrome. Additionally, we did not perform large deletion/duplication analysis of FBN1 that may have impacted our mutation detection rate slightly. On the other hand, 55/207 (27%) of the patients in this cohort whom tested positive for a pathogenic or likely pathogenic FBN1 variant did not fulfill 2010 Ghent criteria. The observation of FBN1 causal variants in Ghent-negative and, on the other hand, lack of FBN1 causal variants in Ghent-positive patients is likely due to the largely heterogenous nature of the disorder as well as the fact that the criteria used to establish MFS are not completely specific or sensitive. Therefore, our data support that FBN1 genetic testing can be beneficial to help substantiate a diagnosis of MFS, and also to provide a basis for potentially ruling out the disorder or considering another, overlapping genetic disorder.
Current practice generally considers that FBN1 missense variants are associated with more classic MFS, and patients with non-missense FBN1 variants may have a milder disease presentation. However, the frequency of missense variants in our Ghent-positive patients was significantly lower than the Ghent-negative patients (41 vs 89%; P<0.0001). We observed a similar lower frequency of missense variants between Ghent-positive and -negative patients in adults and young individuals. As compared with other studies examining variant type in Ghent-positive patients, the frequency of missense variants observed in our cohort (41%) was lower than in previous reports (59%;19 50%;22 and 51%23). However, these previous studies occurred before the 2010 change to the Ghent nosology criteria, which may have impacted the differences observed between our study and the previous studies. Furthermore, the criteria used to establish variant pathogenicity is more robust than in previous years due to increased knowledge about human genetic variation and access to large variant databases. A more recent smaller study of 24 Ghent-positive patients, utilizing Ghent 2010 criteria, observed a 54% missense variant frequency, although a comparison was not made with patients who were Ghent criteria negative.20 Our novel observation that Ghent-positive patients have a lower frequency of missense variants is contrary to current practice (which would anticipate that most Ghent-positive patients would have missense variants). We have observed a similar phenomenon with a lower frequency of missense variants in MFS patients with aortic events.24 Future studies examining mechanism of missense vs non-missense FBN1 variants in pathogenesis of MFS could help to further our understanding of this topic.
FBN1 is a large gene, comprising 65 exons, in which numerous alterations have been previously reported. Here, we add to that body of literature by describing an additional 130 novel FBN1 variants that fall into the categories of pathogenic, likely pathogenic or VUS. In the pathogenic or likely pathogenic variant category (n=207), 109 of these variants were novel. Similar to previous reports, we observed that the majority of FBN1 variants that occurred in our overall cohort were missense. Those missense variants that were pathogenic or likely pathogenic were likely to occur in highly conserved regions of the gene, for example, impacting a critical cysteine residue in a cbEGF-like or TB domain.
Familial studies have been known to be beneficial in helping to elucidate the pathogenicity of novel and previously reported VUSs. In our study, we were able to evaluate family members of 19 probands with novel missense variants and novel or previously reported VUSs. For 11 of these cases, familial testing helped to determine or substantiate variant pathogenicity. There were eight inconclusive cases where either the variant did not track with suspected disease in family members, or not enough information was provided to determine whether the variant tracked with disease, or too few family members were tested to substantiate pathogenicity or lack thereof.
It has been previously suggested that there is a relationship between severity of ocular involvement and the presence of a cysteine substitution or other FBN1 pathogenic variant in specific regions of the gene.2, 6, 17, 25 Consistent with what others found, we observed that individuals with EL were significantly more likely to have missense variants involving a cysteine residue and pathogenic variants in exons 1–15, and were less likely to have truncating variants. We also confirmed previous findings of patients with early onset, rapidly progressing MFS and pathogenic variants in exons 24–32.
In MFS suspected cases where an FBN1 genetic variant is identified, the results may sometimes be difficult to interpret in terms of their impact on protein function, especially if they are novel missense or intronic variants (outside the critical splice junction region). It is well known that interpretation of the clinical significance of genetic variants is often dependent on knowledge of previously reported specific variants, known gene regions/residues critical for protein function, familial studies and genotype–phenotype correlations. Additionally, it is known that MFS can present variably, and is sometimes undiagnosed or unsuspected in patients presenting with isolated features of MFS. Recent strides in large databases containing human genetic variation, such as the EVS, have greatly enhanced our ability to properly classify genetic variants. As we move down the path of increased utilization of large-scale genetic testing, it is likely that scenarios identifying FBN1 variants will increase in frequency, thus substantiating the need to publish new variants with associated clinical data.
With our report, we have added to this body of literature by reporting multiple novel FBN1 variants, as well as familial testing information and phenotypic details in the patients who harbor these variants. A novel observation reported here is the lower frequency of missense variants occurring in patients with positive Ghent criteria status. The implications for this novel observation could be important since it conflicts with the general mindset that patients with non-missense variants have a milder course of disease. Our study additionally points to the imperfect nature of relying solely on clinical criteria to establish a diagnosis of MFS, and the benefit of utilizing genetic testing to aid in diagnosis. Overall, the information detailed here can be utilized as an aid in the clinical interpretation of FBN1 genetic findings and may act as a springboard for further studies of FBN1 and MFS.
References
Judge, D. P. & Dietz, H. C. Marfan's syndrome. Lancet 366, 1965–1976 (2005).
Faivre, L., Collod-Beroud, G., Loeys, B. L., Child, A., Binquet, C., Gautier, E. et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am. J. Hum. Genet. 81, 454–466 (2007).
Gray, J. R., Bridges, A. B., Faed, M. J., Pringle, T., Baines, P., Dean, J. et al. Ascertainment and severity of Marfan syndrome in a Scottish population. J. Med. Genet. 31, 51–54 (1994).
Turner, C. L., Emery, H., Collins, A. L., Howarth, R. J., Yearwood, C. M., Cross, E. et al. Detection of 53 FBN1 mutations (41 novel and 12 recurrent) and genotype-phenotype correlations in 113 unrelated probands referred with Marfan syndrome, or a related fibrillinopathy. Am. J. Med. Genet. A 149A, 161–170 (2009).
Biggin, A., Holman, K., Brett, M., Bennetts, B. & Ades, L. Detection of thirty novel FBN1 mutations in patients with Marfan syndrome or a related fibrillinopathy. Hum. Mutat. 23, 99 (2004).
Comeglio, P., Johnson, P., Arno, G., Brice, G., Evans, A., Aragon-Martin, J. et al. The importance of mutation detection in Marfan syndrome and Marfan-related disorders: report of 193 FBN1 mutations. Hum. Mutat. 28, 928 (2007).
Rommel, K., Karck, M., Haverich, A., von Kodolitsch, Y., Rybczynski, M., Muller, G. et al. Identification of 29 novel and nine recurrent fibrillin-1 (FBN1) mutations and genotype-phenotype correlations in 76 patients with Marfan syndrome. Hum. Mutat. 26, 529–539 (2005).
Loeys, B. L., Dietz, H. C., Braverman, A. C., Callewaert, B. L., De Backer, J., Devereux, R. B. et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 47, 476–485 (2010).
Aalberts, J. J., Thio, C. H., Schuurman, A. G., van Langen, I. M., van der Pol, B. A., van Tintelen, J. P. et al. Diagnostic yield in adults screened at the Marfan outpatient clinic using the 1996 and 2010 Ghent nosologies. Am. J. Med. Genet. A 158A, 982–988 (2012).
Yang, J. H., Han, H., Jang, S. Y., Moon, J. R., Sung, K., Chung, T. Y. et al. A comparison of the Ghent and revised Ghent nosologies for the diagnosis of Marfan syndrome in an adult Korean population. Am. J. Med. Genet. A 158A, 989–995 (2012).
Khau Van Kien, P., Baux, D., Pallares-Ruiz, N., Baudoin, C., Plancke, A., Chassaing, N. et al. Missense mutations of conserved glycine residues in fibrillin-1 highlight a potential subtype of cb-EGF-like domains. Hum. Mutat. 31, E1021–E1042 (2010).
Downing, A. K., Knott, V., Werner, J. M., Cardy, C. M., Campbell, I. D. & Handford, P. A. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85, 597–605 (1996).
Dietz, H. C. & Pyeritz, R. E. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum. Mol. Genet. 4 Spec No 1799–1809 (1995).
Tiecke, F., Katzke, S., Booms, P., Robinson, P. N., Neumann, L., Godfrey, M. et al. Classic, atypically severe and neonatal Marfan syndrome: twelve mutations and genotype-phenotype correlations in FBN1 exons 24-40. Eur. J. Hum. Genet. 9, 13–21 (2001).
Booms, P., Cisler, J., Mathews, K. R., Godfrey, M., Tiecke, F., Kaufmann, U. C. et al. Novel exon skipping mutation in the fibrillin-1 gene: two 'hot spots' for the neonatal Marfan syndrome. Clin. Genet. 55, 110–117 (1999).
Robinson, P. N., Booms, P., Katzke, S., Ladewig, M., Neumann, L., Palz, M. et al. Mutations of FBN1 and genotype-phenotype correlations in Marfan syndrome and related fibrillinopathies. Hum. Mutat. 20, 153–161 (2002).
Comeglio, P., Evans, A. L., Brice, G., Cooling, R. J. & Child, A. H. Identification of FBN1 gene mutations in patients with ectopia lentis and marfanoid habitus. Br. J. Ophthalmol. 86, 1359–1362 (2002).
Rybczynski, M., Bernhardt, A. M., Rehder, U., Fuisting, B., Meiss, L., Voss, U. et al. The spectrum of syndromes and manifestations in individuals screened for suspected Marfan syndrome. Am. J. Med. Genet. A 146A, 3157–3166 (2008).
Loeys, B., Nuytinck, L., Delvaux, I., De Bie, S. & De Paepe, A. Genotype and phenotype analysis of 171 patients referred for molecular study of the fibrillin-1 gene FBN1 because of suspected Marfan syndrome. Arch. Intern. Med 161, 2447–2454 (2001).
Attanasio, M., Pratelli, E., Porciani, M. C., Evangelisti, L., Torricelli, E., Pellicano, G. et al. Dural ectasia and FBN1 mutation screening of 40 patients with Marfan syndrome and related disorders: role of dural ectasia for the diagnosis. Eur. J. Med. Genet. 56, 356–360 (2013).
Sheikhzadeh, S., Kade, C., Keyser, B., Stuhrmann, M., Arslan-Kirchner, M., Rybczynski, M. et al. Analysis of phenotype and genotype information for the diagnosis of Marfan syndrome. Clin. Genet. 82, 240–247 (2012).
Rand-Hendriksen, S., Tjeldhorn, L., Lundby, R., Semb, S. O., Offstad, J., Andersen, K. et al. Search for correlations between FBN1 genotype and complete Ghent phenotype in 44 unrelated Norwegian patients with Marfan syndrome. Am. J. Med. Genet. A 143A, 1968–1977 (2007).
Arbustini, E., Grasso, M., Ansaldi, S., Malattia, C., Pilotto, A., Porcu, E. et al. Identification of sixty-two novel and twelve known FBN1 mutations in eighty-one unrelated probands with Marfan syndrome and other fibrillinopathies. Hum. Mutat. 26, 494 (2005).
Baudhuin, L. M., Kotzer, K. E. & Lagerstedt, S. A. Increased frequency of FBN1 truncating and splicing variants in Marfan syndrome patients with aortic events. Genet. Med. (e-pub ahead of print 7 August 2014; doi:10.1038/gim.2014.91).
Schrijver, I., Liu, W., Brenn, T., Furthmayr, H. & Francke, U. Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am. J. Hum. Genet. 65, 1007–1020 (1999).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Baudhuin, L., Kotzer, K. & Lagerstedt, S. Decreased frequency of FBN1 missense variants in Ghent criteria-positive Marfan syndrome and characterization of novel FBN1 variants. J Hum Genet 60, 241–252 (2015). https://doi.org/10.1038/jhg.2015.10
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.10
This article is cited by
-
Health-related quality of life in children and adolescents with Marfan syndrome or related disorders: a controlled cross-sectional study
Orphanet Journal of Rare Diseases (2024)
-
Variability in gene-based knowledge impacts variant classification: an analysis of FBN1 missense variants in ClinVar
European Journal of Human Genetics (2019)
-
Interpretation of sequence variants of the FBN1 gene: analog or digital? A commentary on decreased frequency of FBN1 missense variants in Ghent criteria-positive Marfan syndrome and characterization of novel FBN1 variants
Journal of Human Genetics (2015)