Abstract
Mucopolysaccharidosis IVA (MPS IVA; Morquio A syndrome) is a lysosomal storage disease caused by deficiency of N-acetylgalactosamine-6-sulfatase (GALNS) and transmitted as an autosomal recessive trait. This is the first systematic mutation screen in Chinese MPS IVA patients. Mutation detections in 24 unrelated Chinese MPS IVA patients were performed by PCR and direct sequencing of exons or the mRNA of GALNS. A total of 42 mutant alleles were identified, belonging to 27 different mutations. Out of the 27 mutations, 16 were novel, including 2 splicing mutations (c.567-1G>T and c.634-1G>A), 2 nonsense mutations (p.W325X and p.Q422X) and 12 missense mutations (p.T88I, p.H142R, p.P163H, p.G168L, p.H236D, p.N289S, p.T312A, p.G316V, p.A324E, p.L366P, p.Q422K and p.F452L). p.G340D was found to be a common mutation in the Chinese MPS IVA patients, accounting for 16.7% of the total number of mutant alleles. The results show that the mutations in Chinese MPS IVA patients are also family specific but have a different mutation spectrum as compared to those of other populations.
Similar content being viewed by others
Introduction
Mucopolysaccharidosis IVA (MPS IVA), also known as Morquio A syndrome (OMIM No. 252300) is an autosomal recessive metabolic disease, caused by the deficiency of N-acetylgalactosamine-6-sulfatase (GALNS; EC 3.1.6.4). GALNS is involved in the stepwise degradation of keratin sulfate and chondroitin-6-sulfate.1, 2 The deficiency of GALNS leads to accumulation of substrates in lysosomes, resulting in cell dysfunction2 and clinical manifestations.
The phenotype of MPS IVA can be classified into three subgroups according to the patient's final height: severe (<120 cm), intermediate (between 120 and 140 cm) and mild (above 140 cm), respectively.3 The patients of the severe type are also characterized by features such as short stature, genu valgum, odontoid dysplasia, protrusion of the chest, kyphoscoliosis, hypermobility of joints and abnormal gait,3 and are mostly below the 90th percentile on the Morquio A growth charts.4 They often have their onsets during infancy, and cannot survive through the second or third decade of life. The patients of the intermediate and mild types, generally called ‘attenuated’ types, have relatively less skeletal involvement and longer life span.
The GALNS gene (Gene ID: 2588) has been mapped to 16q24.3.5 GALNS contains 14 exons,6 transcribed into a 2.3 kb mRNA7 and translated into a protein of 522 residues.8 The mature GALNS has an arylsulfatase domain that is highly conserved within the human sulfatase protein family and among GALNS of different species.8
To date, 156 different mutations have been identified in GALNS, 70% of which are single-nucleotide substitutions (missense/nonsense mutations).9 Various bioinformatics tools have been used to predict whether a missense mutation is pathogenic. A GALNS-specific scoring system, considering both evolutionary conservation and chemical characteristics of a given residue, has been proposed to assist in such predictions. Mutation spectrums have been created in some populations and distinctions were observed.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 Although one-fifth of the world population is composed of Chinese, the mutation spectrum in Chinese MPS IVA patients is not available.
In this study, mutation detections were performed in 24 Chinese MPS IVA patients. A preliminary mutation spectrum in Chinese patients has been proposed from the results.
Materials and methods
Patients
This study was approved by the ethics committee of the Peking Union Medical College.
The 24 MPS IVA patients included in this study were diagnosed between 1999 and 2008. All patients were first clinically diagnosed according to their symptoms and signs, and subsequently confirmed by GALNS enzyme assay (vide infra) in the McKusick-Zhang Center for Genetic Medicine, Peking Union Medical College Hospital.
These patients resided in different cities or counties in 16 provinces of China. The parents of one of the patients, IV_20, were of Han and Dong lineages; the others were of Han origin. Neither consanguineous marriage nor known kinship between families could be revealed by tracing the family tree according to information provided by the patients or their close relatives. In patients under 18 years, preliminary determination of their positions on the Morquio A-specific growth charts4 was made.
GALNS enzyme assay
GALNS activities were assayed according to the method described by Kleijer et al.20 Leukocytes were separated from 2 ml of peripheral blood and lysed by ultrasonication. 4-Methylumbelliferyl-β-D-galactose-6-sulfate (Moscerdam Substrates, Oegstgeest, the Netherlands) was used as the fluorogenic substrate to determine GALNS activity. The activity was expressed as the amount of substrate (nmol) cleaved per h per mg of protein in the cell lysates. The normal range in Chinese controls was found to be 4.57–9.36 nmol h−1 per mg protein (W Zhang, unpublished data, n=50).
Mutation detection
Genomic DNA was extracted from 1 ml peripheral blood using the sodium chloride/chloroform method, or the Lab-Aid 800 magnetic beads nucleic acid extraction system (BioV, Xiamen, China).
Primer pairs GAL_01 to 14 (listed in Table 1) were designed by the online software Primer3 (http://frodo.wi.mit.edu/)21 to amplify each exon with adjacent intronic sequences (for exon 14, only the translated region was amplified).
PCR was conducted in 50 μl volume containing 20 ng genomic DNA, 10 pmol of each primer, 10 nmol dNTPs, 2.5 U rTaq DNA Polymerase (Takara, Liaoning, China), and 25 μl 2 × GC buffer I or II (Takara) as PCR enhancer. The reactions were performed in a T1 thermocycler (Biometra, Göttingen, Germany) with a thermal profile consisting of initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, annealing at temperatures as listed in Table 1 for 30 s, and 72 °C for 45 s, followed by a final extension at 72 °C for 10 min.
PCR products were purified by agarose gel extraction kit, and then used as templates in sequencing reactions. PCR primers in either direction were used as sequencing primers. Sequencing reactions were performed in a T1 thermocycler (Biometra) with BigDye Terminator v.3.1 (Applied Biosystems, Foster City, CA, USA), following the manufacturer's protocol. Sequences were obtained on ABI Prism 3730 Capillary Array Sequencer (Applied Biosystems).
Sequencing chromatograms were aligned with reference sequences (NCBI build 36.1) by Sequencher software version 4.8 (Gene Codes, Ann Arbor, MI, USA).
The transmission of sequence variations found in each patient was inferred by sequencing corresponding exons of his/her parents. Analysis of novel sequence variations was performed by comparing with the direct sequencing results of corresponding exons in 50 healthy unrelated Chinese Han individuals.
RT–PCR
Total RNA was isolated from 500 μl fresh peripheral blood with TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's protocol.
Reverse transcription (RT) reactions were performed within 24 h after RNA isolation, in a volume of 10 μl containing 100 ng RNA, 50 pmol oligo (dT)18 primer, 5 nmol dNTPs, 10 U RNase inhibitor (Takara), 50 U MMLV reverse transcriptase (Takara) and corresponding reaction buffer. The reaction conditions were as described in the manufacturer's protocol.
Primer pair GAL_RT2, covering a segment from nt 403 to nt 851 of the full-length cDNA (NM_000512.4), were designed by Primer Premier software package version 5.0 (Premier Biosoft, Palo Alto, CA, USA). The procedures of amplification, purification and sequencing were the same as that described above.
Bioinformatics analysis
SIFT and PolyPhen are in silico tools that predict impact of amino-acid substitutions. SIFT predicts whether an amino-acid substitution affects protein function based on sequence homology and the physical properties of amino acids.22 PloyPhen makes predictions of possible impact of an amino-acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations.23 Missense mutations were submitted to SIFT (http://sift.jcvi.org/) and PolyPhen (http://genetics.bwh.harvard.edu/pph/), respectively. Parameters were reserved as default. The GALNS-specific scoring system24 was also used. GALNS protein sequences of 11 species (human, dog, mouse, rat, pig, cattle, puffer fish, frog, chicken, zebrafish and sea urchin) were aligned by Clustal-X2.25 Evolutionary conservation (CEVO) of a given residue was scored as 1, 2, 3 or 4, according to the level of conservation (conserved in all species, conserved in vertebrates, conserved in mammals and not conserved).
Conservativeness of amino-acid substitution (CAAS) was scored as 1, 2 or 3 according to the chemical difference (χ, defined by Grantham26) of the two residues involved in a substitution: nonconservative (χ>90), semiconservative (60<χ⩽90) and conservative (χ⩽60).
The sum (S) of CEVO and CAAS was used in judging the pathogenicity of a missense substitution. Cutoff index between pathogenic and polymorphic was set as 4 as described by Tomatsu et al.24
Haplotype analysis
Up to 34 single-nucleotide polymorphisms (SNPs), including 15 HapMap SNPs, were genotyped for mutation detection. By comparing SNP genotypes of a patient to those of his/her parents, haplotypes of alleles were constructed. SNPs used were rs11862754, rs11865929, rs35137494, rs13334220, rs71395332, rs34278797, rs8059282, rs7196835, rs8054994, rs3743544, rs3743545, rs7187889, rs2269333, rs17603837, rs12934499, rs34745339, rs1064315, rs11076721, rs61742258, rs35232749, rs7187783, rs8050636, rs7191220, rs3743546, rs3833041, rs74035850, rs12446069, rs3859024, rs73251099, rs2303269, rs2303270, rs73251097, rs73251084, rs2303271 (listed in genomic sequence; HapMap SNPs were written in bold).
Results
Clinical features and GALNS activity
Basic information, major clinical features and GALNS activity of each patient were retraced from reserved medical records and listed in Table 2.
Skeletal abnormalities such as pectus carinatum (91.7%, 22/24 patients), short trunk dwarfism (79.2%, 19/24 patients), genu valgum (62.5%, 15/24 patients) and wrist joint laxity (62.5%, 15/24 patients) were the most common findings in these patients.
Patients' GALNS activities ranged from 0 to 0.182 nmol h−1 per mg protein. In most of the patients, it was below 0.050 nmol h−1 per mg protein (62.5%, 15/24 patients), between 0.050 and 0.100 in 5 (20.8%, 5/24 patients), and above 0.100 in 4 (16.7%, 4/24 patients).
In this study, patient IV_12 was 144 cm tall, thus belonging to the mild group. Patients IV_15 (129 cm) and IV_20 (133 cm) were of the intermediate group. Patients still in childhood were classified according to the Morquio A growth chart for each gender.4 Two girls, IV_04 and IV_06, were classified as of the attenuated type; the others were classified as of the severe group.
Mutation detection
A total of 42 mutant alleles were identified in the 24 patients (see Supplementary information) while 6 alleles remained unknown (Table 2). The identified mutant alleles were of 27 different kinds (Table 3), including 1 small deletion, 2 nonsense mutations, 3 splicing mutations and 21 missense mutations. Of these variations, 11 were known mutations and 16 were novel (p.T88I, p.H142R, p.P163H, p.G168L, p.H236D, p.N289S, p.T312A, p.G316V, p.A324E, p.L366P, p.Q422K, p.F452L, p.W325X, p.Q422X, c.567-1G>T, c.634-1G>A). All the novel mutations were not detected in 100 control chromosomes.
Parental origins of most alleles were inferred (Table 3). Heterozygous variations were detected in two adjacent nucleotides (c.502G>T and c.503G>T) on sequencing exon 5 of patient IV_24. The two variations probably existed in cis on the maternal allele for they were both transmitted from the patient's mother.
RT–PCR
RNA of patients IV_15 and IV_24 was extracted from peripheral blood to validate effects of splicing mutations through RT–CR.
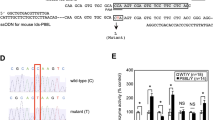
Patient IV_15 was a heterozygote of c.567-1G>T and p.P163H. As shown in Figure 1, overlapping peaks, starting from nt 205 on the sequencing chromatogram, were found to be due to the superposition of a normal allele and an aberrant allele with an 11 bp deletion (r.567_577del).
In patient IV_24, overlapping peaks started from coding DNA nts 423 (Figure 2). The overlapping peaks were found to be due to the superposition of a normal allele and an aberrant allele with a 144 bp deletion (r.423_566del) and with skipping of exon 5.
Bioinformatics analysis
Twenty-one missense mutations (12 novel and 9 known mutations) identified in this study were evaluated. Effects of missense mutations predicted by PolyPhen, SIFT and the GALNS-specific methods as well as CEVO, CAAS and S were listed in Table 3.
All mutations were presumed to be pathogenic by at least one method. Of the 21 mutations, 11 were so classified by all three methods, 6 by two methods and 4 by one method.
In 16 of the 21 mutations, the affected residue is conserved in all species investigated, from sea urchin to human (CEVO=1). Three mutations (p.G168L, p.L366P and p.Q422K) were conserved in vertebrates (CEVO=2), and two mutations (p.F452L and p.G340D) were conserved in mammals (CEVO=3). Of the 21 missense mutations, 12 were conservative (CAAS=3) or semiconservative (CAAS=2), and 9 were nonconservative (CAAS=1).
Haplotype analysis
A missense mutation c.1019G>A (p.G340D) was detected in eight mutant alleles of five patients. Haplotype analysis was performed to examine whether the eight p.G340D-alleles descended from the same ancestor.
Genotypes were identical for 33 SNPs (including all 15 HapMap SNPs) in all eight alleles. The only exception was rs35137494 located in exon 2: in two p.G340D-alleles cytosine was present, whereas in the other six, thymine was present.
Discussion
This study was carried out with two purposes: (1) to detect mutations in the Chinese MPS IVA patients and to provide prenatal diagnosis to affected families if desired, and (2) to work out a preliminary mutation spectrum of the Chinese patients, and compare it with those of the global populations.
Of the 27 mutations, 16 identified in this study were novel, corresponding to 10% of the present GALNS mutation database (http://www.hgmd.org).
As a member of the sulfatase protein family, GALNS functions through a conserved sulfatase domain, composed of codon 31–453 (http://pfam.sanger.ac.uk/protein?acc=P34059). The 3D structural model of human GALNS has been constructed by homology modeling of the X-ray crystal structure of human 4S and ASA,27 although the X-ray crystal structure of GALNS was not available.
Both novel nonsense mutations, p.W325X and p.Q422X, may result in a defective arylsulfatase domain. p.W325X could cause the loss of some N-terminal components (the last β-sheet and last two α-helixes) and all C-terminal components (four β-sheets and one α-helix); p.Q422X would lead to the loss of the last C-terminal α-helix.24, 27
Two novel intronic splicing mutations, c.567-1G>T and c.634-1G>A, were the first splicing defects identified in introns 5 and 6. Both mutations affected the 3′-acceptor site, which is believed to be the most conserved intronic splicing elements. Mutation in the 3′-acceptor site usually lead to false splicing, resulting in marked structural change of the resulting enzyme protein.
The consequence of c.567-1G>T was disclosed by sequencing analysis of RT–PCR product of patient IV_15, which resulted in a frameshift with premature termination at codon 193. The 11-bp deletion in mRNA (r.567_577del) revealed the existence of a cryptic splicing acceptor, hidden 11 bp downstream of the beginning of exon 6. The cryptic splicing acceptor would be activated when the normal splicing acceptor was disrupted. This supports the fact that effects of mutations in conserved intronic splicing elements could be different from retention of intron(s), or skipping of exon(s). Local structures should be carefully scrutinized when making predictions.
r.423_566del was discovered by RT–PCR, resulting in the formation of an immediate stop codon (TGA, TG from exon 4 and A from exon 6). The genomic cause of this mutation is still unclear. All exons of the patient were sequenced and no splicing mutation was found. Additional investigation is required to disclose the genomic change responsible for this mutation.
Predicting effects of single-nucleotide variations in coding region were considered to be an easy task by checking the table of genetic code. It is actually more complicated. Exonic single-nucleotide variations in GALNS may act in one or more of the following ways: (1) as a polymorphism that does not affect the gene's functions, (2) as a missense mutation resulting in reduced enzyme activities and (3) as a missense mutation that affects the formation of lysosomal multienzyme complex which is responsible for the stabilization of GALNS.24, 28, 29
In vitro expression of mutant alleles has been used to judge the pathogenicity of missense mutations,30 by assaying enzyme activities of plasmid containing mutant cDNA produced by site-direct mutagenesis. This method may be limited in distinguishing substitutions of residues involved in forming lysosomal multienzyme complex if they do not change enzyme activities greatly. Other methods, such as bioinformatics tools with distinct design and algorithm, may help to improve the prediction.
Three bioinformatics tools for missense mutations are based on different designs. PloyPhen is based on multiple sequence alignment and protein 3D structures; SIFT is based on sequence homology among proteins similar in sequence and function, either the same protein in different species or other members of the same protein family; the GALNS-specific scoring system considers conservation of residues among aligned GALNSs in species at different evolutionary level (CEVO) and conservativeness of chemical characteristics between substituted amino acids (CAAS).
Generally, predictions for mutations in the N-terminus of GALNS have been more consistent using all three softwares than predictions for mutations in the C-terminus. This may be because results of multi-alignment of sequence similar to GALNS are considered in algorithms of all three methods, whereas the conserved sulfatase domain is mainly located in the N-terminus. However, aligned sequences were different in the three methods. Only GALNS sequences of different species are used in the GALNS-specific method, while sequences aligned in SIFT and PolyPhen include sequences of other proteins or less reliable sequences similar to those of GALNS. Inclusion of less-reliable sequences may produce confusions in the results and reduce the quality of prediction.
This is the first systematic mutation analysis in Chinese MPS IVA patients. Both mutations previously reported in Taiwan Chinese patients18 (p.M318R and p.L36_L37del) were included in our study.
Comparing with the mutations reported previously in global populations, 17 out of 27 (or 63%) of the mutations found in Chinese patients (16 novel mutations described in this study and p.L36_L37del described previously in a Taiwan Chinese patient) are not observed in other nations so far, indicating a different mutation spectrum may exist in Chinese population.
Eight p.G340D-alleles were detected in five Chinese patients (three homozygotes and two compound heterozygotes). It is the most common mutant allele in the Chinese population, accounting for 16.7% (8/48) of the total mutant alleles among Chinese. All 5 patients with p.G340D were residents or emigrants from adjacent provinces in central eastern China (Hebei, Shandong, Anhui and Henan) within a circumference of 600 km. These eight alleles had identical haplotype in formed by 33 of 34 SNPs in exon 2–13. The only difference was rs35137494, with cytosine in two and thymine in six of the eight p.G340D alleles. rs35137494 is located in a CpG dinucleotide, which has been proved to have a higher mutation rate due to its biochemical structure. Considering the above information, it is more likely that p.G340D in these patients could be identical-by-descent and derived from the same founder. The only difference in the haplotypes could be caused by a recurrent transition of rs35137494. Therefore, similar to the situations previously described in Italian17 and Finnish31 patients, the mutation spectrum in Chinese patients is unique because most mutations are Chinese-unique and family-specific, whereas one mutation (p.G340D) exists in a higher proportion (16.7%). The unique mutation spectrum is in agreement with the fact that Chinese were comparatively isolated genetically in history due to its geographic location and culture.
In this study, 87.5% (42/48) of the mutant alleles were characterized. Six mutant alleles remain unknown. Similarly, in other mutation detection programs for MPS IVA,10, 11, 12, 13, 14, 15, 19 there were also about 10–25% alleles undetected. This may be due to the limitation of the exon-by-exon-sequencing method currently used, which detects only mutations in discontinuous short amplicons. Chromosomal rearrangements cannot be detected unless cytogenetic method is applied. Heterozygotes with deletion or duplication covering the entire PCR amplicon(s)32 may be missed, because sequencing data from one single allele or from two same alleles cannot be differentiated. RNA analysis or quantitative methods, such as real-time PCR or multiplex ligation-dependent Probe amplification, may be of help in such cases.
Single-nucleotide mutations may also be missed. Untranslated regions are usually less focused on in routine mutation screening programs; however, some mutations in untranslated region have been reported in LSDs.33 A number of nucleotides in the genome, coding or noncoding, have been found to function in the regulation of transcription, splicing and translation. Characteristics of these cis-acting regulatory elements are not clearly understood and therefore such mutations may be misjudged as missense mutation or synonymous polymorphism34, 35 or left in the unchecked region of introns36 or interval between genes. More efforts, such as RNA-based strategy, are required to detect these unknown alleles for better understanding of GALNS gene mutations and for more precise prenatal gene diagnosis.
As a recessive disease, the clinical phenotype is manifested only when both alleles are not functioning. Linkage phase of detected variants should be confirmed to avoid the situation when more variations in one allele are present. Errors in gene diagnosis will end up in a wrong prenatal diagnosis.
In conclusion, this is the first systematic mutation screening program in Chinese MPS IVA patients. In this study, we identified 16 novel mutations in GALNS, corresponding to 10% of the mutation database. p.G340D was found to be a common mutation in the Chinese population. These findings can be of benefit to MPS IVA patients in gene diagnosis, drug discovery and therapy development. As we mentioned, in certain cases linkage phase of mutations, RNA analysis and bioinformatics tools can be very valuable.
References
Di Ferrante, N., Ginsberg, L. C., Donnelly, P. V., Di Ferrante, D. T. & Caskey, C. T. Deficiencies of glucosamine-6-sulfate or galactosamine-6-sulfate sulfatases are responsible for different mucopolysaccharidoses. Science 199, 79–81 (1978).
Singh, J., Di Ferrante, N., Niebes, P. & Tavella, D. N-acetylgalactosamine-6-sulfate sulfatase in man. Absence of the enzyme in Morquio disease. J. Clin. Invest. 57, 1036–1040 (1976).
Montano, A. M., Tomatsu, S., Gottesman, G. S., Smith, M. & Orii, T. International Morquio A Registry: clinical manifestation and natural course of Morquio A disease. J. Inherit. Metab. Dis. 30, 165–174 (2007).
Montano, A. M., Tomatsu, S., Brusius, A., Smith, M. & Orii, T. Growth charts for patients affected with Morquio A disease. Am. J. Med. Genet. A 146A, 1286–1295 (2008).
Masuno, M., Tomatsu, S., Nakashima, Y., Hori, T., Fukuda, S., Masue, M. et al. Mucopolysaccharidosis IV A: assignment of the human N-acetylgalactosamine-6-sulfate sulfatase (GALNS) gene to chromosome 16q24. Genomics 16, 777–778 (1993).
Morris, C. P., Guo, X. H., Apostolou, S., Hopwood, J. J. & Scott, H. S. Morquio A syndrome: cloning, sequence, and structure of the human N-acetylgalactosamine 6-sulfatase (GALNS) gene. Genomics 22, 652–654 (1994).
Nakashima, Y., Tomatsu, S., Hori, T., Fukuda, S., Sukegawa, K., Kondo, N. et al. Mucopolysaccharidosis IV A: molecular cloning of the human N-acetylgalactosamine-6-sulfatase gene (GALNS) and analysis of the 5′-flanking region. Genomics 20, 99–104 (1994).
Tomatsu, S., Fukuda, S., Masue, M., Sukegawa, K., Fukao, T., Yamagishi, A. et al. Morquio disease: isolation, characterization and expression of full-length cDNA for human N-acetylgalactosamine-6-sulfate sulfatase. Biochem. Biophys. Res. Commun. 181, 677–683 (1991).
Tomatsu, S., Montano, A. M., Nishioka, T., Gutierrez, M. A., Pena, O. M., Tranda Firescu, G. G. et al. Mutation and polymorphism spectrum of the GALNS gene in mucopolysaccharidosis IVA (Morquio A). Hum. Mutat. 26, 500–512 (2005).
Tomatsu, S., Dieter, T., Schwartz, I. V., Sarmient, P., Giugliani, R., Barrera, L. A. et al. Identification of a common mutation in mucopolysaccharidosis IVA: correlation among genotype, phenotype, and keratan sulfate. J. Hum. Genet. 49, 490–494 (2004).
Tomatsu, S., Nishioka, T., Montano, A. M., Gutierrez, M. A., Pena, O. S., Orii, K. O. et al. Mucopolysaccharidosis IVA: identification of mutations and methylation study in GALNS gene. J. Med. Genet. 41, e98 (2004).
Terzioglu, M., Tokatli, A., Coskun, T. & Emre, S. Molecular analysis of Turkish mucopolysaccharidosis IVA (Morquio A) patients: identification of novel mutations in the N-acetylgalactosamine-6-sulfate sulfatase (GALNS) gene. Hum. Mutat. 20, 477–478 (2002).
Tomatsu, S., Fukuda, S., Cooper, A., Wraith, J. E., Ferreira, P., Di Natale, P. et al. Fourteen novel mucopolysaccharidosis IVA producing mutations in GALNS gene. Hum. Mutat. 10, 368–375 (1997).
Kato, Z., Fukuda, S., Tomatsu, S., Vega, H., Yasunaga, T., Yamagishi, A. et al. A novel common missense mutation G301C in the N-acetylgalactosamine-6-sulfate sulfatase gene in mucopolysaccharidosis IVA. Hum. Genet. 101, 97–101 (1997).
Bunge, S., Kleijer, W. J., Tylki-Szymanska, A., Steglich, C., Beck, M., Tomatsu, S. et al. Identification of 31 novel mutations in the N-acetylgalactosamine-6-sulfatase gene reveals excessive allelic heterogeneity among patients with Morquio A syndrome. Hum. Mutat. 10, 223–232 (1997).
Laradi, S., Tukel, T., Khediri, S., Shabbeer, J., Erazo, M., Chkioua, L. et al. Mucopolysaccharidosis type IV: N-acetylgalactosamine-6-sulfatase mutations in Tunisian patients. Mol. Genet. Metab. 87, 213–218 (2006).
Tomatsu, S., Filocamo, M., Orii, K. O., Sly, W. S., Gutierrez, M. A., Nishioka, T. et al. Mucopolysaccharidosis IVA (Morquio A): identification of novel common mutations in the N-acetylgalactosamine-6-sulfate sulfatase (GALNS) gene in Italian patients. Hum. Mutat. 24, 187–188 (2004).
Yang, C. F., Tsai, F. J., Lin, S. P., Lee, C. C. & Wu, J. Y. A novel in-frame deletion mutation (c106-111del) identified in a Taiwan Chinese patient with type IVA mucopolysaccharidosis. Hum. Mutat. 18, 254 (2001).
Yamada, N., Fukuda, S., Tomatsu, S., Muller, V., Hopwood, J. J., Nelson, J. et al. Molecular heterogeneity in mucopolysaccharidosis IVA in Australia and Northern Ireland: nine novel mutations including T312S, a common allele that confers a mild phenotype. Hum. Mutat. 11, 202–208 (1998).
Kleijer, W. J., Geilen, G. C., Garritsen, V., Huijmans, J. G., Los, F. J., Voznyi, Y. V. et al. First-trimester diagnosis of Morquio disease type A. Prenat. Diagn. 20, 183–185 (2000).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Sunyaev, S., Ramensky, V., Koch, I., Lathe, W. III, Kondrashov, A. S. & Bork, P. Prediction of deleterious human alleles. Hum. Mol. Genet. 10, 591–597 (2001).
Tomatsu, S., Montano, A. M., Lopez, P., Trandafirescu, G., Gutierrez, M. A., Oikawa, H. et al. Determinant factors of spectrum of missense variants in mucopolysaccharidosis IVA gene. Mol. Genet. Metab. 89, 139–149 (2006).
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Grantham, R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974).
Sukegawa, K., Nakamura, H., Kato, Z., Tomatsu, S., Montano, A. M., Fukao, T. et al. Biochemical and structural analysis of missense mutations in N-acetylgalactosamine-6-sulfate sulfatase causing mucopolysaccharidosis IVA phenotypes. Hum. Mol. Genet. 9, 1283–1290 (2000).
Ostrowska, H., Krukowska, K., Kalinowska, J., Orlowska, M. & Lengiewicz, I. Lysosomal high molecular weight multienzyme complex. Cell Mol. Biol. Lett. 8, 19–24 (2003).
Pshezhetsky, A. V. & Ashmarina, M. Lysosomal multienzyme complex: biochemistry, genetics, and molecular pathophysiology. Prog. Nucleic Acid. Res. Mol. Biol. 69, 81–114 (2001).
Fukuda, S., Tomatsu, S., Masue, M., Sukegawa, K., Iwata, H., Ogawa, T. et al. Mucopolysaccharidosis type IVA. N-acetylgalactosamine-6-sulfate sulfatase exonic point mutations in classical Morquio and mild cases. J. Clin. Invest. 90, 1049–1053 (1992).
Montano, A. M., Kaitila, I., Sukegawa, K., Tomatsu, S., Kato, Z., Nakamura, H. et al. Mucopolysaccharidosis IVA: characterization of a common mutation found in Finnish patients with attenuated phenotype. Hum. Genet. 113, 162–169 (2003).
Fukuda, S., Tomatsu, S., Masuno, M., Ogawa, T., Yamagishi, A., Rezvi, G. M. et al. Mucopolysaccharidosis IVA: submicroscopic deletion of 16q24.3 and a novel R386C mutation of N-acetylgalactosamine-6-sulfate sulfatase gene in a classical Morquio disease. Hum. Mutat. 7, 123–134 (1996).
Gieselmann, V., Polten, A., Kreysing, J. & von Figura, K. Arylsulfatase A pseudodeficiency: loss of a polyadenylylation signal and N-glycosylation site. Proc. Natl Acad. Sci. USA 86, 9436–9440 (1989).
Wu, Y., Zhang, Y. & Zhang, J. Distribution of exonic splicing enhancer elements in human genes. Genomics 86, 329–336 (2005).
Wang, J., Smith, P. J., Krainer, A. R. & Zhang, M. Q. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 33, 5053–5062 (2005).
Davis, R. L., Homer, V. M., George, P. M. & Brennan, S. O. A deep intronic mutation in FGB creates a consensus exonic splicing enhancer motif that results in afibrinogenemia caused by aberrant mRNA splicing, which can be corrected in vitro with antisense oligonucleotide treatment. Hum. Mutat. 30, 221–227 (2009).
Acknowledgements
We thank all patients and their family members who participated in this study. This work was supported by grants from Beijing Science and Technology Committee Foundation (D0906005040491) and the National Supporting Program of the 11th Five-year Plan Period for Science and Technology (2006BAI05A08).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Wang, Z., Zhang, W., Wang, Y. et al. Mucopolysaccharidosis IVA mutations in Chinese patients: 16 novel mutations. J Hum Genet 55, 534–540 (2010). https://doi.org/10.1038/jhg.2010.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.65
Keywords
This article is cited by
-
Clinical and genetic characteristics of concomitant Mucopolysaccharidosis type IVA and neurogenic bladder in children: two case reports and literature review
BMC Pediatrics (2021)
-
Abnormally increased carotid intima media-thickness and elasticity in patients with Morquio A disease
Orphanet Journal of Rare Diseases (2020)
-
Clinical, biochemical and genetic profiles of patients with mucopolysaccharidosis type IVA (Morquio A syndrome) in Malaysia: the first national natural history cohort study
Orphanet Journal of Rare Diseases (2019)
-
Demographic characteristics and distribution of lysosomal storage disorder subtypes in Eastern China
Journal of Human Genetics (2016)