Abstract
16q-ADCA (OMIM no. 117210) is an autosomal dominant spinocerebellar ataxia (AD-SCA) characterized by late-onset pure cerebellar ataxia and −16C>T substitution of the puratrophin-1 gene. Recently, a series of single-nucleotide polymorphisms (haplotype block) were found to be specific to 16q-ADCA. We screened patients with ataxia and found 62 patients, including four homozygotes who carry the C–T substitution of the puratrophin-1 gene. By further analysis of the patients with the haplotype block, we observed a single-founder effect for 16q-ADCA, even in patients who are supposed to be sporadic late cortical cerebellar atrophy (LCCA). We also observed slippage mutations of microsatellite markers, GATA01 and 17msm, in the pedigrees. We compared the clinical course of 16q-ADCA in heterozygotes and homozygotes with the haplotype block and observed no apparent gene dosage effect. 16q-ADCA accounts for 27% of AD-SCAs and is the most frequent AD-SCA in South Kyushu, Japan.
Similar content being viewed by others
Introduction
Autosomal dominant cerebellar ataxias (ADCAs) are clinically classified into three subtypes, and ADCA type III was defined as pure cerebellar ataxia with late onset.1 Until date, 16 genes and 10 additional loci are reported to cause ADCAs.2, 3 SCA4 belongs to ADCA type I, characterized by cerebellar ataxia with sensory axonal neuropathy.4 Its locus was originally mapped to 16q22.1 in a family from Utah and was confirmed in SCA4 families from Germany.4, 5 The 16q-ADCA from six Japanese families was mapped to the same region as SCA4;6 however, it was categorized as ADCA type III on the basis of clinical features.6, 7, 8 A single-nucleotide substitution (−16C>T) in the 5′-untranslated region of the puratrophin-1 gene is strongly associated with 16q-ADCA.9 However, three patients from families having 16q-ADCA (−16C>T) did not have the substitution.10, 11, 12 Haplotype analysis suggested that the mutation was in a region centromeric to the −16C>T substitution.10, 11, 12
We had reported that this disease locus was telomeric to the puratrophin-1 gene.8 To clarify the disease locus in these patients, we reexamined the disease haplotype using microsatellite markers and a series of single-nucleotide polymorphisms (SNPs) specific to 16q-ADCA. Moreover, we reported the clinical features of 49 patients, including four homozygous patients, and described the frequency of 16q-ADCA in South Kyushu, Japan.
Materials and methods
We studied 147 patients with a family history of ataxia and 331 patients with sporadic ataxias, referred to our department (Department of Neurology and Geriatrics, Kagoshima University Graduate School of Medical and Dental Sciences) from South Kyushu (Kagoshima, Miyazaki and Oita) and Okinawa Island during the past decade until January 2008. All patients were referred by their primary physicians or neurologists and signed informed consents were obtained from the patients. The study was approved by the Institutional Review Board of Kagoshima University. Genomic DNA was extracted from peripheral blood leukocytes. We screened for the expansion of triplet repeats associated with SCAs (SCA1, -2, -3, -6, -7, -8, SCA12, -17 and DRPLA) by standard PCR methods and GeneScan analysis software using an ABI Prism 377 DNA sequencer (Foster City, CA, USA) as described earlier.8 We used PCR-direct sequencing or PCR-restriction fragment length polymorphism to examine the C–T substitution of puratrophin-1 and the Gerstmann–Sträussler–Scheinker syndrome (GSS) mutation.9, 13 For patients who had a C–T substitution of the puratrophin-1 gene, PCR product sizes of the microsatellite markers, GATA01 and 17msm, were determined using the ABI Prism 377 Genetic Analyzer, (Foster City, CA, USA).8 Using PCR-direct sequencing, we typed SNP02, -04, -05 and -06, which are highly specific to the diseased chromosome.12 The SCA5, SCA10, SCA14 and FGF mutations were not analyzed in this study. Expert neurologists provided information on the clinical features of the 16q-ADCA patients. The Kaplan–Meier method was used to draw the curve of clinical severity for puratrophin-1 alteration of 43 heterozygotes and four homozygotes, with the end point being wheelchair bound. The log-rank test was used for comparison between the two groups because of the small number of homozygotes. Patient data were statistically processed using StatView version 5.0.
Results
Of the 478 patients, 62 carried the puratrophin-1 C–T substitution. Of the 62 affected individuals, 43 patients were from 17 families with an autosomal dominant inheritance. Eleven patients were from seven families who had no proof of autosomal dominant inheritance, but their siblings had similar neurological symptoms. Eight patients had either sporadic ataxias or unclear family histories. We examined all the patients with puratrophin-1 alteration to determine whether they had a haplotype block (SNP05 and SNP06, highly specific to the 16q-ADCA allele), but neither of the SNPs was identified in 200 control chromosomes.12 All patients had changes in SNP05 (G–A) and SNP06 (A–G), whereas none of the 96 controls had the haplotype block (Table 1).
The average age of onset of ataxia for the 49 patients with the C–T change was 59 years (range, 43–80 years), and the oldest asymptomatic carrier (K01 pedigree) was 80 years old (Table 2).
The initial symptoms of ADCA are gait ataxia or dysarthria. On neurological examination, all individuals were affected with truncal and limb ataxia. Although deep-tendon reflexes were generally increased, spasticities or pathological reflexes were rare. Hearing impairment and dementia were poorly represented in our cohort. Brain magnetic resonance imaging revealed that 97% of the affected individuals had cerebellar atrophy, especially in the vermis.
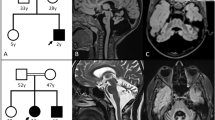
We compared the clinical features between the four homozygous and 45 heterozygous patients. One homozygous patient had a clear family history, and both his parents, who were maternal/paternal cousins of each other, had medical histories of ataxia. Three siblings homozygous for the substitution were found by our genetic analysis. The mother of these patients reported a history of ataxia, but their father did not have ataxic symptoms and died of old age at 82 years. Similar to the heterozygous patients, the mean age of the four homozygous patients at the time of onset was 59.3 years (range, 50–63 years). However, within each family, the disease onset of the homozygotes tended to be earlier than that of the heterozygotes. The duration of the period until being wheelchair bound was determined for four homozygotes and 43 heterozygotes by the Kaplan–Meier method. There was no clinical difference between the two groups (P=0.9689) (Figure 1).
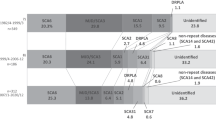
The results of the family-based study in 116 families diagnosed by gene tests for all patients revealed that 16q-ADCA (27%) was the most prevalent ADCA in South Kyushu and Okinawa, followed by SCA6 (21%), MJD/SCA3 (15%), DRPLA (9%) and GSS (9%). The results of the patient-based study also revealed that 16q-ADCA was the most prevalent ADCA. The 16q-ADCA accounted for 65% of the ADCAs in Miyazaki Prefecture and was the most frequent ADCA in Kagoshima Prefecture (Figure 2). Characteristically, GSS accounted for 17.5% of ADCAs in Kagoshima Prefecture. In Oita Prefecture, SCA6 was the most frequent ADCA, followed by 16q-ADCA. DRPLA and SCA2 consisted mostly of ADCAs, and 16q-ADCA was not found in Okinawa Prefecture.
Relative frequency of ADCAs in the prefectures of South Kyushu, Japan. For Kagoshima, patients were referred from the whole area of Kagoshima, hence the cohort represents Kagoshima Prefecture. On the other hand, the data for Miyazaki, Oita and Okinawa prefectures were collected from a limited number of hospitals; therefore, these data may not represent the exact frequency of the disease in each prefecture.
Discussion
The 16q-ADCA has been mapped to 16q22.1.6, 7, 8 It has been reported that the −16C>T substitution of the puratrophin-1 gene makes it possible to genetically diagnose 16q-ADCA.9 16q-ADCA families have been reported from various parts of Japan after the discovery of puratrophin-1 alteration, and it is one of the most common ataxic diseases in Japan.10, 11, 14, 15, 16, 17 Although the pathogenicity of the −16C>T substitution is controversial because of the discovery of two patients from families having 16q-ADCA, who did not have the −16C>T substitution,10, 11 it remains useful for the screening of 16q-ADCA in Japanese patients, as the −16C>T substitution has never been seen in Japanese controls thus far.12
We found 32 pedigrees within 62 affected individuals who carried the −16C>T substitution of puratrophin-1. Nineteen patients were believed to be recessive for SCA or sporadic LCCA, but we revealed that all 19 had the haplotype block (SNP05-SNP06-−16C>T substitution of puratrophin-1), which is highly specific to 16q-ADCA. This indicates that all of our 16q-ADCA patients originated from a single founder. Therefore, these sporadic patients are not de novo in origin, but their diagnoses might be because of limited ancestral information. 16q-ADCA patients diagnosed by the puratrophin-1 alteration among sporadic LCCA have also been reported in earlier studies.14, 15, 16 We agree with their proposal that screening for the −16C>T substitution of puratrophin-1 should be considered for SCA diagnosis, even in cases believed to be sporadic LCCA.14, 16
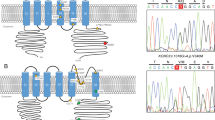
Earlier, we excluded a region centromeric to 17msm as critical for 16q-ADCA, because, compared with other pedigrees, patients with the K01 pedigree have a differently sized marker at GATA01 and 17msm on both sides of the −16C>T substitution.8 Nevertheless, the K01 pedigree has the haplotype block specific to 16q-ADCA. The disease haplotype of pedigree K01 is A-G-162-T-189 at SNP05-SNP06-GATA01-puratrophin-1-17msm. Conversely, most pedigrees have the haplotype, A-G-158-T-191. If the human genome sequence is correct in this critical region, the difference in microsatellite marker size is interpretable as being either multiple recombinations among SNP06, GATA01, puratrophin-1 and 17msm, or a slippage mutation of the microsatellite markers; however, multiple recombinations are less likely to be based on the limited distance (approximately 0.75 Mb) from SNP06 to 17msm. As there is only a single-repeat unit difference between GATA01 and 17msm, a slippage mutation might have occurred. Microsatellite slippage mutations in GATA01, D16S397 and GGAA10 were also reported in 16q-ADCA families.12 When the point mutation rate was 10−8 per nucleotide per generation, the slippage mutation rates ranged from 10−7 to 10−2.18 It might be better to use SNPs for 16q-ADCA mapping rather than microsatellite markers, as there are three disease-specific SNPs, which suggests a greater occurrence of slippage mutations around the SNPs. On the basis of haplotype analysis with these SNPs, we reassign SNP04 to its centromeric border and the puratrophin-1 alteration to its telomeric border. The chromosomal position should be from 64 982 677 to 65 871 433 on chromosome 16 of the human genome sequence (NCBI Build 36.1) (Figure 3).
In agreement with other studies, the clinical features of our 16q-ADCA patients were late-onset pure cerebellar ataxia and brisk deep-tendon reflex.11, 14, 15, 16, 17 We found that hearing impairment, dementia and leukoaraiosis varied on the basis of patient location, suggesting that environmental factors or genetic backgrounds other than puratrophin-1 mutations are responsible for 16q-ADCA symptoms.
In an earlier study, four patients from one pedigree showed homozygosity for the puratrophin-1 C–T substitution, and their mean age at onset was earlier than that of heterozygotes within the same pedigree.14 In this study, we found four homozygotes from two pedigrees and compared their phenotypic severity with that of heterozygotes over several years. We did not find differences between homozygotes and heterozygotes with regard to age at onset or phenotypic severity. The four homozygotes had the A-G-T haplotype at SNP05-SNP06-puratrophin-1, indicating homozygosity for the 16q-ADCA mutation. The gene dosage effect for AD-SCAs varies by gene. If the mechanism of 16q-ADCA is haploinsufficiency caused by the total or partial loss of function of a single allele, then a homozygote should show a severe phenotype. As we found no homozygotic patients with severe phenotypes, this suggests an alternative mechanism. Homozygotes with triplet-repeat expansions of SCA2, SCA3 or SCA6 showed modified or severe phenotypes.19, 20, 21, 22, 23, 24 Conversely, homozygotes of SCA3, SCA6, SCA8 and SCA12 did not have severe clinical phenotypes compared with heterozygotes.19, 25, 26, 27, 28 On the basis of our results, there does not seem to be a strong gene dosage effect in 16q-ADCA. As some homozygotes with SCAs caused by gain-of-function mutations did not always show as severe a phenotype as the heterozygote, it is reasonable to suggest that 16q-ADCA operates through a gain-of-function mechanism.
The 16q-ADCA prevalence in Miyazaki and Kagoshima districts was higher than the average prevalence in Japan. The GSS syndrome (Prion protein (PRNP) Pro102Leu mutation) accounted for 18% of the AD-SCAs in Kagoshima Prefecture. Although the inclusion of GSS as an AD-SCA might be controversial, we included it on the basis of prominent cerebellar ataxia that has led physicians to recognize it as an ADCA. This epidemiological study should help neurologists gain an understanding of the frequency of ADCAs in the district.
References
Harding, A. E. Clinical features and classification of inherited ataxias. Adv. Neurol. 61, 1–14 (1993).
Schols, L., Bauer, P., Schmidt, T., Schulte, T. & Riess, O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 3, 291–304 (2004).
Duenas, A. M., Goold, R. & Giunti, P. Molecular pathogenesis of spinocerebellar ataxias. Brain 129, 1357–1370 (2006).
Flanigan, K., Gardner, K., Alderson, K., Galster, B., Otterud, B., Leppert, M. F. et al. Autosomal dominant spinocerebellar ataxia with sensory axonal neuropathy (SCA4): clinical description and genetic localization to chromosome 16q22.1. Am. J. Hum. Genet. 59, 392–399 (1996).
Hellenbroich, Y., Bubel, S., Pawlack, H., Opitz, S., Vieregge, P., Schwinger, E. et al. Refinement of the spinocerebellar ataxia type 4 locus in a large German family and exclusion of CAG repeat expansions in this region. J. Neurol. 250, 668–671 (2003).
Nagaoka, U., Takashima, M., Ishikawa, K., Yoshizawa, K., Yoshizawa, T., Ishikawa, M. et al. A gene on SCA4 locus causes dominantly inherited pure cerebellar ataxia. Neurology 54, 1971–1975 (2000).
Li, M., Ishikawa, K., Toru, S., Tomimitsu, H., Takashima, M., Goto, J. et al. Physical map and haplotype analysis of 16q-linked autosomal dominant cerebellar ataxia (ADCA) type III in Japan. J. Hum. Genet. 48, 111–118 (2003).
Hirano, R., Takashima, H., Okubo, R., Tajima, K., Okamoto, Y., Ishida, S. et al. Fine mapping of 16q-linked autosomal dominant cerebellar ataxia type III in Japanese families. Neurogenetics 5, 215–221 (2004).
Ishikawa, K., Toru, S., Tsunemi, T., Li, M., Kobayashi, K., Yokota, T. et al. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am. J. Hum. Genet. 77, 280–296 (2005).
Ohata, T., Yoshida, K., Sakai, H., Hamanoue, H., Mizuguchi, T., Shimizu, Y. et al. A −16C>T substitution in the 5′ UTR of the puratrophin-1 gene is prevalent in autosomal dominant cerebellar ataxia in Nagano. J. Hum. Genet. 51, 461–466 (2006).
Nozaki, H., Ikeuchi, T., Kawakami, A., Kimura, A., Koide, R., Tsuchiya, M. et al. Clinical and genetic characterizations of 16q-linked autosomal dominant spinocerebellar ataxia (AD-SCA) and frequency analysis of AD-SCA in the Japanese population. Mov. Disord. 22, 857–862 (2007).
Amino, T., Ishikawa, K., Toru, S., Ishiguro, T., Sato, N., Tsunemi, T. et al. Redefining the disease locus of 16q22.1-linked autosomal dominant cerebellar ataxia. J. Hum. Genet. 52, 643–649 (2007).
Petraroli, R., Vaccari, G. & Pocchiari, M. A rapid and efficient method for the detection of point mutations of the human prion protein gene (PRNP) by direct sequencing. J. Neurosci. Methods 99, 59–63 (2000).
Ouyang, Y., Sakoe, K., Shimazaki, H., Namekawa, M., Ogawa, T., Ando, Y. et al. 16q-linked autosomal dominant cerebellar ataxia: a clinical and genetic study. J. Neurol. Sci. 247, 180–186 (2006).
Onodera, Y., Aoki, M., Mizuno, H., Warita, H., Shiga, Y. & Itoyama, Y. Clinical features of chromosome 16q22.1 linked autosomal dominant cerebellar ataxia in Japanese. Neurology 67, 1300–1302 (2006).
Hayashi, M., Adachi, Y., Mori, M., Nakano, T. & Nakashima, K. Clinical and genetic epidemiological study of 16q22.1-linked autosomal dominant cerebellar ataxia in western Japan. Acta. Neurol. Scand. 116, 123–127 (2007).
Basri, R., Yabe, I., Soma, H. & Sasaki, H. Spectrum and prevalence of autosomal dominant spinocerebellar ataxia in Hokkaido, the northern island of Japan: a study of 113 Japanese families. J. Hum. Genet. 52, 848–855 (2007).
Lai, Y. & Sun, F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20, 2123–2131 (2003).
Lerer, I., Merims, D., Abeliovich, D., Zlotogora, J. & Gadoth, N. Machado–Joseph disease: correlation between the clinical features, the CAG repeat length and homozygosity for the mutation. Eur. J. Hum. Genet. 4, 3–7 (1996).
Geschwind, D. H., Perlman, S., Figueroa, K. P., Karrim, J., Baloh, R. W. & Pulst, S. M. Spinocerebellar ataxia type 6. Frequency of the mutation and genotype–phenotype correlations. Neurology 49, 1247–1251 (1997).
Ikeuchi, T., Takano, H., Koide, R., Horikawa, Y., Honma, Y., Onishi, Y. et al. Spinocerebellar ataxia type 6: CAG repeat expansion in alpha1A voltage-dependent calcium channel gene and clinical variations in Japanese population. Ann. Neurol. 42, 879–884 (1997).
Sasaki, H., Wakisaka, A., Sanpei, K., Takano, H., Igarashi, S., Ikeuchi, T. et al. Phenotype variation correlates with CAG repeat length in SCA2—a study of 28 Japanese patients. J. Neurol. Sci. 159, 202–208 (1998).
Ragothaman, M., Sarangmath, N., Chaudhary, S., Khare, V., Mittal, U., Sharma, S. et al. Complex phenotypes in an Indian family with homozygous SCA2 mutations. Ann. Neurol. 55, 130–133 (2004).
Ragothaman, M. & Muthane, U. Homozygous SCA 2 mutations changes phenotype and hastens progression. Mov. Disord. 23, 770–771 (2008).
Takiyama, Y., Sakoe, K., Namekawa, M., Soutome, M., Esumi, E., Ogawa, T. et al. A Japanese family with spinocerebellar ataxia type 6 which includes three individuals homozygous for an expanded CAG repeat in the SCA6/CACNL1A4 gene. J. Neurol. Sci. 158, 141–147 (1998).
Tazon, B., Badenas, C., Jimenez, L., Munoz, E. & Mila, M. SCA8 in the Spanish population including one homozygous patient. Clin. Genet. 62, 404–409 (2002).
Izumi, Y., Maruyama, H., Oda, M., Morino, H., Okada, T., Ito, H. et al. SCA8 repeat expansion: large CTA/CTG repeat alleles are more common in ataxic patients, including those with SCA6. Am. J. Hum. Genet. 72, 704–709 (2003).
Bahl, S., Virdi, K., Mittal, U., Sachdeva, M. P., Kalla, A. K., Holmes, S. E. et al. Evidence of a common founder for SCA12 in the Indian population. Ann. Hum. Genet. 69, 528–534 (2005).
Acknowledgements
We thank Ms A Yoshimura of Kagoshima University for her excellent technical assistance. We also thank Dr Chihaya Koriyama, an associate professor of Epidemiology and Preventive Medicine of Kagoshima University, for her help on statistical analyses. This study was supported in part by grants from the Nervous and Mental Disorders and Research Committee for Ataxic Disease of the Japanese Ministry of Health, Welfare and Labor (Grant 19A-1; HT and KA), Frontier Science Research Center Program (FSRC) of Kagoshima University (HT), and the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant 19591001; HT).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hirano, R., Takashima, H., Okubo, R. et al. Clinical and genetic characterization of 16q-linked autosomal dominant spinocerebellar ataxia in South Kyushu, Japan. J Hum Genet 54, 377–381 (2009). https://doi.org/10.1038/jhg.2009.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.44
Keywords
This article is cited by
-
Prevalence of Fragile X-Associated Tremor/Ataxia Syndrome in Patients with Cerebellar Ataxia in Japan
The Cerebellum (2021)
-
Eye movement changes in autosomal dominant spinocerebellar ataxias
Neurological Sciences (2020)
-
Autosomal dominant cerebellar ataxia type III: a review of the phenotypic and genotypic characteristics
Orphanet Journal of Rare Diseases (2013)