Abstract
The LMNA gene was sequenced in 98 Czech patients from 94 unrelated families with early-onset axonal Charcot–Marie–Tooth (CMT) disease consistent with both autosomal recessive inheritance and sporadic cases. Biallelic pathogenic mutations were not found in any patient in this group. One patient carried the c.1870C>T mutation that is predicted to result in the amino-acid substitution, p. Arg624Cys, on one allele, but the second causative mutation was not detected. LMNA mutation is not likely to be associated with the disease in this family. To exclude larger deletions/duplications in the LMNA gene not detectable by sequencing, 48 patients from this group were also analyzed with multiplex ligation-dependent probe amplification. No rearrangements in the LMNA gene were detected. We conclude that mutations in the LMNA gene are absent from a large group of Czech patients with axonal autosomal recessive CMT disease. Consequently, LMNA mutation screening does not seem to be relevant for axonal CMT DNA diagnostics. A similar situation may apply to other European populations.
Similar content being viewed by others
Introduction
Charcot–Marie–Tooth (CMT) disease or hereditary motor and sensory neuropathy is traditionally classified according to clinical, electrophysiological, morphological and genetic criteria.1, 2, 3 CMT disorders are divided into two groups on the basis of nerve conduction studies—median motor nerve conduction velocity (MNCV): (1) demyelinating types (CMT1) with MNCV <38 m s−1; and (2) axonal types (CMT2) with MNCV >38 m s−11.
In addition to the neurophysiological features of axonal neuropathy, axonal CMT disease is also characterized histopathologically by chronic axonal atrophy and regeneration in the nerve biopsy. Clinically, the main symptoms in 90% of the cases are early onset, symmetrical muscle weakness and wasting (predominantly in the distal lower limbs), foot deformities and walking difficulties associated with reduced or absent tendon reflexes.4, 5 In autosomal recessive axonal CMT (AR CMT) associated with mutation of lamin A/C,6, 7 weakness and amyotrophy of the upper limbs and an involvement of the proximal muscles of the lower limbs are frequent.
To date, three loci have been associated with AR CMT28: 1q21.2–21.3 for AR CMT2A or CMT2B19; 19q13.3 for CMT2B210; and 8q21.3 for CMT2G.11 Specific mutations in two genes have been identified: ganglioside-induced differentiation protein 1 (GDAP1)12 and lamin (LMNA).13
Mutations in the LMNA gene that encodes the proteins of the inner nuclear membrane, lamin A and C, are involved in at least seven distinct genetic disorders.14 Cases displaying a combined phenotype of these entities are also described.15
Mutations in the LMNA gene are associated with axonal AR CMT,13 but have also been found in a family presenting an autosomal dominant mode of inheritance of CMT2 in combination with muscular dystrophy, cardiomyopathy and leuconychia.16
For several reasons, the LMNA gene was considered as a good candidate gene for axonal CMT.13 First, lamin A/C is a member of the intermediate filament family, and the other intermediate filament protein, NEFL, is already involved in CMT2E.17 Second, lamin A/C expression patterns during neuronal development were found to be important for neurons.18 Observations of transgenic LMNA-null mice gave further support to this assumption.13, 19
Currently, it is unknown how frequent mutations are in the LMNA gene among patients worldwide with axonal CMT. Our study was designed to determine the frequency and spectrum of mutations in the LMNA gene among Czech patients with axonal AR CMT and to evaluate the relevance of the LMNA gene for further diagnostic DNA testing in CMT2 patients.
Materials and methods
Patients
Charcot–Marie–Tooth (CMT) disease or hereditary neuropathy is a heterogeneous group of disorders and is the most frequent hereditary neuromuscular disorder with a frequency of 1:2500 in the population.20 We would expect the Czech population, consisting of 10 million, to have 4000 CMT patients. As our laboratory is the only one in the Czech Republic testing for CMT, all patient samples are collected in our registry and our data can provide evidence for the incidence of CMT in the Czech Republic. To date, DNA samples from almost 3000 patients suspected of having CMT from about 1500 unrelated families have been collected in our laboratory. CMT has been confirmed by finding a causal mutation in 1077 individuals of Czech origin. A CMT1A duplication was detected in 521 individuals from 282 unrelated families, an HNPP deletion was detected in 302 individuals from 170 families and a mutation in other CMT-related genes was detected in 254 individuals from 102 families.
A total of 98 patients from 94 unrelated Czech axonal CMT families were screened for mutations in the LMNA gene, by direct sequencing of all 12 coding exons and exon/intron boundaries.
Inclusion criteria were the following:
-
1)
All selected families were compatible with autosomal recessive inheritance, that is, the patients had affected siblings and healthy parents based on family history (four families, eight patients), or more often sporadic cases (90 patients). Consanguinity was not reported for any of the families.
-
2)
Only patients with axonal neuropathy (MNCV >38 m s−1) were included.
-
3)
Most patients presented with first neuropathy symptoms early in life (first or second decade).
In 13.3% (13 patients), symptoms started in the first 5 years of life, 16.3% of patients (16) presented with the first symptoms between ages 5–10 years. In 14.3% (14) of patients, age at onset was during the second decade of life. The disease started in the third decade in 4.1% of patients (4). Few patients (8.1%, eight patients) were older than 30 years at disease onset. Forty-three patients presented with first symptoms of axonal CMT disease in the first or second decade, but precise age is unknown (43.9%).
Most patients showed symmetrical muscle weakness in the lower limbs and foot deformities. Five patients complained about weakness of the arms, six about paresthesias and three patients are wheel-chair bound.
In 74 patients, deletions/duplications in the 17p11.2 region were excluded earlier. Only a few patients were tested for mutations in MPZ- (24 patients), MFN2- (16 patients) and GDAP1 (23 patients)-gene with negative results.
Forty-eight patients who showed no heterozyosity for intragenic LMNA polymorphisms were then tested with the P048 multiplex ligation-dependent probe amplification (MLPA) analysis kit from MRC-Holland (Amsterdam, The Netherlands).
All tested patients granted informed consent and the study was approved by the Central Ethical Committee of the University Hospital Motol Prague.
Genetic study
Direct sequencing
DNA was extracted from peripheral blood or from saliva. All 12 exons and exon/intron boundaries of the LMNA gene were PCR amplified in 12 fragments with the primer set 1-1F/1-1R, 1-2F/1-2R, 2F-2R, 3F-3R, 4F-4R, 5F-5R, 6F-6R, 7F-7R, 8+9F-8+9R, 10F-10R, 11F-11R and 12F-12R (Table 1: LMNA Primers set). PCR conditions are listed in Table 2. Purified PCR products (Agencourt Ampure, Beckman Coulter, Fullerton, CA, USA) were sequenced using four dye terminator chemistry (BigDye Terminator v.3.1, Applied Biosystems, Foster City, CA, USA). All sequencing reactions were carried out under conditions recommended by the manufacturer. After purification with the CleanSeq kit (Beckman Coulter), sequencing products were analyzed on an ABI3100-Avant automated genetic analyzer (Applied Biosystems). Sequence traces were compared with an LMNA reference sequence (NM_170707, http://www.ncbi.nlm.nih.gov/sites/entrez) using Sequencing analysis software (Applied Biosystems) and SeqMan software (DNASTAR, Madison, WI, USA).
Multiplex ligation-dependent probe amplification analysis method
Forty-eight patients were tested by MLPA analysis (MRC Holland) to exclude larger deletions/duplications in the LMNA gene (www.mlpa.com). The SALSA MLPA kit (MRC-Holland), P048 LMNA, was used (lot 1106, 0106). This kit contains probes for 10 of the 12 coding exons of the LMNA gene and is designed to detect deletions/ duplications of one or more exons of the gene. MLPA analysis was carried out according to the manufacturer's protocol (http://www.mlpa.com/pages/support_mlpa_protocolspag.html). Capillary electrophoresis of the MLPA-PCR products used either an ABI 310 or ABI3100-Avant genetic analyzer (Applied Biosystems). GeneMapper software v.4.0, GeneScan v.3.7 and Peak Scanner software (all Applied Biosystems) were used to extract data. The results were evaluated both visually and statistically. For visual evaluation, the recommendation of the manufacturer was followed. Statistical analysis of the results was carried out using Excel sheets prepared by A Wallace, NGRL (Manchester), UK (www.ngrl.org.uk). Analysis spreadsheets are available upon request. For autosomal loci, a deletion was indicated by a relative reduction of 35–55% in the peak area for that probe amplification product, and a duplication was indicated by a relative gain in the peak area between 30 and 50%.
Results
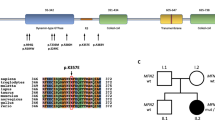
Biallelic pathogenic mutations in the LMNA gene were not detected in any of the 98 Czech axonal CMT patients. In one patient with early-onset axonal CMT, we found a novel mutation in exon 11, c.1870C>T (p.Arg624Cys), in the heterozygous state. A second causative mutation was not found in this patient even after MLPA analysis. A healthy father and a healthy brother of the patient also carried this mutation (c.1870 C>T) in the heterozygous state.
We detected several SNP variants. All polymorphisms were detected in a heterozygous state. The polymorphisms identified in this study are listed in Table 3.
No rearrangements (duplications/deletions) in the LMNA gene were found on MLPA analysis in any of the 48 Czech patients with axonal CMT.
Discussion
In this study, we did not find pathogenic biallelic defects at the nucleotide sequence level of the LMNA gene among Czech patients with axonal AR CMT. Moreover, we did not identify any deletions/duplications by MLPA analysis among a subgroup of 48 patients who were selected as those more likely to carry deletions. These results showed that defects in the LMNA gene are either absent, or are extremely rare in Czech patients with axonal CMT. Therefore, we cannot recommend LMNA analysis for diagnostic testing of Czech patients with axonal CMT.
In one patient with severe early-onset axonal CMT, the missense mutation, p.Arg624Cys, was detected. This mutation is present in a heterozygous state in the affected as well as in healthy family members. This mutation is unlikely to be the cause of CMT disease in this family, but it may be pathogenic. However, this mutation is localized in exon 11 of the LMNA gene, and because of alternative splicing, exon 11 is transcribed in Lamin A but not in Lamin C. Therefore, it is not a core part of the LMNA protein. This could explain the nonpathogenic effect of this amino-acid substitution.
To exclude the possibility of a frequent polymorphism, DNA samples from healthy controls were tested for the presence of c.1870C>T. It was not present in any of the 110 healthy chromosomes tested. We concluded that p.Arg624Cys is a rare polymorphism. However, the significance of this mutation would need to be resolved by an additional study at the RNA level.
Our results are consistent with other studies, as few patients with a mutation in the LMNA gene have been described to date in the literature. Bouhouche et al.9 was the first to describe that LMNA is a possible candidate gene for axonal AR CMT in his study on a large consanguineous Moroccan family. Later, several Algerian families were shown to carry a p.Arg298Cys mutation in the LMNA gene.6 A linkage study for three of these families suggested a common ancestral haplotype and gave evidence of a founder effect.13 In 2007, four Moroccan families were presented with mutation p.Arg298Cys in the LMNA gene.8 All CMT2B1 patients, homozygous for this mutation, originate from a restricted region of north-west Algeria and eastern Morocco and carry a homozygous common ancestral haplotype at the LMNA locus.21 Two other mutations in the LMNA gene have been identified in patients with peripheral neuropathy, but always in combination with other clinical signs.15, 16 Recently, authors from China published results similar to ours with the absence of LMNA mutations in 32 sporadic CMT2 patients.22
In conclusion, mutations in the LMNA gene are not a significant cause of axonal Charcot–Marie–Tooth disease in Czech patients. This may also apply in other European countries or even worldwide, because there are few other reports of LMNA mutations in AR CMT2 patients.
References
Dyck, P. J. & Lambert, E. H. Lower motor and sensory neuron diseases with peroneal muscular atrophy. I. Neurologic, genetic, and electrophysiologic findings in hereditary polyneuropathies. Arch. Neurol. 18, 603–618 (1968).
Harding, A. E. & Thomas, P. K. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 103, 259–280 (1980).
Reilly, M. M. Classification of the hereditary motor and sensory neuropathies. Curr. Opin. Neurol. 13, 561–564 (2000).
Gemignani, F. & Marbini, A. Charcot-Marie-Tooth disease (CMT): distinctive phenotypic and genotypic features in CMT type 2. J. Neurol. Sci. 184, 1–9 (2001).
Jonghe, P. D., Timmerman, V. & Broeckhoven, C. V. 2nd Workshop of the European CMT Consortium: 53rd ENMC International Workshop on Classification and Diagnostic Guidelines for Charcot-Marie-Tooth Type 2 (CMT2– HMSN II) and Distal Hereditary Motor Neuropathy (Distal HMN–Spinal CMT), September 26–28, 1997, Naarden, The Netherlands. Neuromuscul. Disord. 8, 426–431 (1998).
Chaouch, M., Allal, Y., De Sandre-Giovannoli, A., Vallat, J. M., Amer-el-Khedoud, A., Kassouri, N. et al. The phenotypic manifestations of autosomal recessive axonal Charcot-Marie-Tooth due to a mutation in Lamin A/C gene. Neuromuscul. Disord. 3, 60–67 (2003).
Tazir, M., Azzedine, H., Assami, S., Sindou, P., Nouioua, S., Zemmouri, R. et al. Phenotypic variability in autosomal recessive axonal Charcot-Marie-Tooth disease due to the R298C mutation in lamin A/C. Brain. 127, 154–163 (2004).
Bouhouche, A., Birouk, N., Azzedine, H., Benomar, A., Durosier, G., Ente, D. et al. Autosomal recessive axonal Charcot-Marie-Tooth disease (ARCMT2): phenotype- genotype correlations in 13 Moroccan families. Brain. 130, 1062–1075 (2007).
Bouhouche, A., Benomar, A., Birouk, N., Mularoni, A., Meggouh, F., Tassin, J. et al. A locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 1q21.2-q21.3. Am. J. Hum. Genet. 65, 722–727 (1999).
Leal, A., Morera, B., Del Valle, G., Heuss, D., Kayser, C., Berghoff, M. et al. A second locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 19q13.3. Am. J. Hum. Genet. 68, 269–274 (2001).
Barhoumi, C., Amouri, R., Ben Hamida, C., Ben Hamida, M., Machghoul, S., Gueddiche, M. et al. Linkage of a new locus for autosomal recessive axonal form of Charcot-Marie-Tooth disease to chromosome 8q21.3. Neuromuscul. Disord. 11, 27–34 (2001).
Baránková, L., Vyhnálková, E., Züchner, S., Mazanec, R., Sakmaryova, I., Vondracek, P. et al. GDAP1 mutations in Czech families with early-onset CMT. Neuromuscul. Disord. 17, 482–489 (2007).
De Sandre-Giovannoli, A., Chaouch, M., Kozlov, S., Vallat, J. M., Tazir, M., Kassouri, N. et al. Homozygous defects in LMNA, encoding lamin A/C nuclear- envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth type 2) and mouse. Am. J. Hum. Genet. 70, 726–736 (2002).
Worman, H. J. & Courvalin, J. C. How do mutations in lamins A and C cause disease? J. Clin. Invest. 113, 349–351 (2004).
Benedetti, S., Bertini, E., Iannaccone, S., Angelini, C., Trisciani, M., Toniolo, D. et al. Dominant LMNA mutations can cause combined muscular dystrophy and peripheral neuropathy. J. Neurol. Neurosurg. Psychiatr. 76, 1019–1021 (2005).
Goizet, C., Yaou, R. B., Demay, L., Richard, P., Bouillot, S., Rouanet, M. et al. A new mutaton of the lamin A/C gene leading to autosomal dominant axonal neuropathy, muscular dystrophy, cardiac disease, and leuconychia. J. Med. Genet. 41, e29 (2004).
Mersiyanova, I. V., Perepelov, A. V., Polyakov, A. V., Sitnikov, V. F., Dadali, E. L., Oparin, R. B. et al. A new variant of Charcot-Marie-Tooth disease type 2 is propably the result of a mutation in the neurofilament-light gene. Am. J. Hum. Genet. 67, 37–46 (2000).
Pierce, T., Worman, H. J. & Holy, J. Neuronal differentiation of NT2/D1 teratocarcinoma cells is accompanied by a loss of lamin A/C expression and an increase in lamin B1 expression. Exp. Neurol. 157, 241–250 (1999).
Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K. et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920 (1999).
Kochanski, A. Molecular genetics studies in Polish Charcot-Marie-Tooth families. Folia Neuropathol. 43, 65–73 (2005).
Hamadouche, T., Poitelon, Y., Genin, E., Chaouch, M., Tazir, M., Kassouri, N. et al. Founder effect and estimation of the age of the c.892C>T (p.Arg298Cys) mutation in LMNA associated to Charcot-Marie-Tooth subtype CMT2B1 in families from North Western Africa. Ann. Hum. Genet. 72 (Pt 5), 590–597 (2008).
Song, S. J., Zhang, Y. Z., Chen, B., Wang, MJ., Wang, Y. Y., Zhang, Y. J. et al. No mutation was detected in the LMNA gene among sporadic Charcot-Marie-Tooth patients. Beijing Da Xue Xue Bao 38, 78–79 (2006).
Acknowledgements
This study was supported by the Czech Ministry of Health (Grant IGA NR 8330-3). We thank our patients and their families for their participation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Laššuthová, P., Baránková, L., Haberlová, J. et al. Mutations in the LMNA gene do not cause axonal CMT in Czech patients. J Hum Genet 54, 365–368 (2009). https://doi.org/10.1038/jhg.2009.43
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.43