Abstract
EGR2 (early growth response 2) is a crucial transcription factor for the myelination of the peripheral nervous system. Mutations in EGR2 are reported to cause a heterogenous spectrum of peripheral neuropathy with wide variation in both severity and age of onset, including demyelinating and axonal forms of Charcot-Marie Tooth (CMT) neuropathy, Dejerine-Sottas neuropathy (DSN/CMT3), and congenital hypomyelinating neuropathy (CHN/CMT4E). Here we report a sporadic de novo EGR2 variant, c.1232A > G (NM_000399.5), causing a missense p.Asp411Gly substitution and discovered through whole-exome sequencing (WES) of the proband. The resultant phenotype is severe demyelinating DSN with onset at two years of age, confirmed through nerve biopsy and electrophysiological examination. In silico analyses showed that the Asp411 residue is evolutionarily conserved, and the p.Asp411Gly variant was predicted to be deleterious by multiple in silico analyses. A luciferase-based reporter assay confirmed the reduced ability of p.Asp411Gly EGR2 to activate a PMP22 (peripheral myelin protein 22) enhancer element compared to wild-type EGR2. This study adds further support to the heterogeneity of EGR2-related peripheral neuropathies and provides strong functional evidence for the pathogenicity of the p.Asp411Gly EGR2 variant.
Similar content being viewed by others
Introduction
Charcot-Marie-Tooth (CMT) neuropathy is group of degenerative motor and sensory peripheral neuropathies which are clinically and genetically heterogenous. Pathogenic variants in over 90 genes cause CMT, and whole-exome sequencing (WES) is now an effective tool for screening known causative genes in unsolved CMT families. Pathogenic variants in the EGR2 gene (early growth response 2) cause a broad spectrum of peripheral neuropathy phenotypes. This includes two forms of severe early-onset peripheral neuropathy, Dejerine-Sottas neuropathy (DSN/CMT3)1,2,3 and congenital hypomyelinating neuropathy (CHN/CMT4E)4,5, as well as adult-onset demyelinating CMT1D with mild-moderate symptoms3,4,5,6,7,8,9,10,11,12 and variable-onset axonal CMT with varied symptom severity13,14.
EGR2 encodes a C2H2-type zinc-finger transcription factor that regulates the expression of genes involved in the formation and maintenance of myelin, including GJB1 (gap junction beta 1), MPZ (myelin protein zero), MBP (myelin basic protein), MAG (myelin associated glycoprotein), PRX (periaxin), and PMP22 (peripheral myelin protein 22)4,15,16,17,18. Approximately half of the reported disease-associated EGR2 variants are de novo and are associated with a severe phenotype13. Here, we report a sporadic de novo EGR2 variant in the third zinc-finger domain, c.1232A > G p.Asp411Gly, discovered through WES candidate gene screening. This variant manifested as a severe DSN phenotype with early onset at two years of age.
Results
Clinical history and neurological examination
The proband is a 56-year-old male who was diagnosed with DSN at six years of age following a nerve biopsy (results not available). He had no family history of peripheral neuropathy, and no family consanguinity. His father and mother had normal nerve conduction studies (NCS) in their fifties. His eldest sister also had a normal neurological examination and NCS at age 25, and the proband reported that this sister has not developed symptoms of peripheral neuropathy in her fifties.
At two years of age, his mother noted that he was consistently fatigued and was repeatedly falling when attempting to walk. He also reported loss of sensation in his feet in early childhood and was never able to run. He was not a candidate for ankle-foot orthoses, and subsequently had a triple arthrodesis at age 15 to stabilise his left ankle. He was also diagnosed with scoliosis and had a Harrington rod inserted at age 16 to correct this. During his teenage years, he had multiple unexplained episodes of hemiparesis and double vision after severe migraine pain behind his eyes, and these episodes would last for approximately two weeks. He reports his mother and sister also suffered from frequent severe migraines, particularly between the ages of 30–45.
On examination at the age of 31, he presented with muscle wasting of the hands and feet, as well as pes cavus, hammer toes, corns, and callouses. He walked with a high steppage gait. He had mildly weak hip flexion, and mildly weak ankle dorsiflexion (3/5). He had absent reflexes in the knees and ankles, with distal loss of vibration sense to the knees. There was loss of pain sensation to above the ankles and decreased joint position sense in the toes. He had a tremor of the hands, with weakness of wrist and finger extension. NCS conducted when the patient was 36 years old (Table 1) showed a severe demyelinating motor and sensory neuropathy.
Autonomic testing conducted at this time showed no postural hypotension, normal cardiovascular reflexes, and normal sudomotor reflexes in both upper and lower limbs indicating no significant autonomic neuropathy. An audiogram showed mild bilateral hearing loss above 1500 Hz, and he also reported occasional tinnitus. Right and left ear brainstem auditory evoked potential (BAEP) waveforms were poorly formed with a bilateral delay in wave V. Right and left full and half field visual evoked potentials (VEP) showed delayed P100 waveforms. An MRI of the brain showed multiple subcortical and periventricular white matter hyperintensities on T2 and FLAIR sequences.
On re-examination at age 52, NCS of the left median sensory, left median motor, and left peroneal motor nerves showed no response. He also reported difficulty opposing his thumb and fingers, and a progressive loss of sensation in his right hand. On examination, there was slight curling of the fingers and wasting of the intrinsic hand muscle. Dorsiflexion in his fingers was mildly weak (grade 3/5), as was his finger abduction (grade 3/5). An additional NCS conducted 12 months later additionally showed no response of the left ulnar sensory nerve, and a severely reduced latency of 19.5 m/s in the left ulnar motor nerve. Four years later at the age of 56, his ability to oppose his thumb and fingers had worsened, and he reported that he was unable to write, or use a knife and fork. He additionally had allodynia below his knees. Another brain MRI was conducted at this time (Fig. 1), which again showed multiple bilateral inactive demyelination foci of T2/FLAIR white matter intensity which were mostly periventricular, however they were also present subcortically (mostly in the frontal lobes), at the callososeptal interface and in the right thalamus. There was no restricted diffusion or suspicious enhancement, and the findings were similar to those from an MRI performed age 37 years. A lumbar puncture for oligoclonal bands was not performed.
Whole-exome sequencing reveals a de novo EGR2 c. 1232A > G missense variant
Microsatellite marker analysis confirmed the parental paternity of both the proband and his sister (Supplementary Fig. 1). Testing for the PMP22 duplication and variants in GJB1 and MPZ were both negative. Pathogenic expansions related to SCA1 (ATXN1), SCA2 (ATXN2), SCA3 (ATXN3), SCA6 (CACNA1A), SCA7 (ATXN7), SCA12 (PPP2R2B), and SCA17 (TBP) were also excluded.
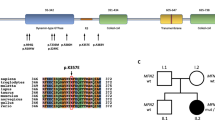
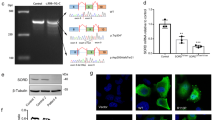
Whole-exome sequencing (WES) was performed on the proband (II:2). Variant filtering of WES was performed as previously described19 and four non-synonymous variants were identified in genes associated with inherited peripheral neuropathy. One variant in the EGR2 gene (NM_000399.5:c.1232A > G) was not previously reported in variant databases including NCBI dbSNP20, 1000 Genomes21, gnomAD22, and ExAC23. Three additional variants were also considered given their low minor allele frequency (MAF <0.1%) (CACNA1A, c.6692G > A, NM_023035.2; BAG3 c.494C > T, NM_004281.3; and ITPR1 c.6692A > G, NM_001168272.1). Sanger sequencing confirmed the heterozygous EGR2 c.1232A > G variant in the proband (Fig. 2a), which was absent in the unaffected mother, father, and sister (Fig. 2b). The variants CACNA1A c.6692G > A (NM_023035.2), BAG3 c.494C > T (NM_004281.3), and ITPR1 c.6692A > G (NM_001168272.1) did not segregate with the peripheral neuropathy phenotype. These results confirmed that the proband is heterozygous for a previously unreported de novo variant in the third zinc-finger domain of EGR2 [chr10: 62,813,406T > C (hg38)], leading to the amino acid substitution p.Asp411Gly.
(a) Sequencing traces of the variant c.1232A > G EGR2 in the affected proband (II:2) and the wild-type sequence in the unaffected father (I:1). Genbank sequence NM_000399.3 was used as a reference for the EGR2 coding sequence. (b) Pedigree of the two-generation kindred and associated EGR2 c.1232A > G genotypes. Solid square denotes the affected male, an open square denotes an unaffected male, and an open circle denotes an unaffected female. An asterisk indicates individuals sent for Sanger sequencing of the EGR2 c.1232A > G variant.
Due to the additional phenotype of hemiplegic migraine in our proband, as well as migraine in his mother and eldest sister, variant filtering of WES was also conducted for genes related to familial hemiplegic migraine (FMH). As variants in CACNA1A can also cause an FMH phenotype in addition to an inherited neuropathy, segregation of the CACNA1A c.6692G > A variant was conducted. However, it was present in the mother and absent in the FMH-affected eldest sister. Additionally, the proband had no variants in the ATP1A2 or SCN1A genes, which are both known to cause FMH.
In silico analyses show that p.Asp411Gly EGR2 affects a highly conserved residue and that it is likely pathogenic
Amino acid sequence alignment demonstrates that the p.Asp411Gly variant occurs at a highly conserved residue in EGR2 with surrounding amino-acid residues being conserved between orthologues in different vertebrate species (Fig. 3). Multiple in silico analysis techniques were utilised to predict the conservation of the Asp411 amino-acid residue and pathogenicity of the p.Asp411Gly variant. GERP24, phastCons25, and PhyloP26 scores all provided further support that this amino-acid residue is highly evolutionarily constrained (Table 2). The functional effect of the p.Asp411Gly variant was predicted to be damaging by Polyphen227, PROVEAN28, SIFT29, and MutationTaster230 (Table 2).
p.Asp411Gly EGR2 has reduced transcriptional regulatory activity compared to wild-type EGR2 in cultured schwann cells
To determine the relevant functional consequences of the p.Asp411Gly EGR2 variant, the ability of wild-type and p.Asp411Gly EGR2 to induce activity of a previously reported EGR2 response element at the PMP22 locus was tested in a Schwann cell line31. A construct expressing wild-type or p.Asp411Gly EGR2 was transfected into cultured rat Schwann (S16) cells along with a construct harbouring an EGR2 response element upstream of a minimal promoter32 and a firefly luciferase (Fluc) reporter gene. To control for cell viability and transfection efficiency, a pCMV-Renilla luciferase (Rluc) construct was also transfected. The fold induction of Fluc, measured as the ratio of Fluc activity to Rluc activity, was employed to test the ability of EGR2 (wild-type or p.Asp411Gly) to activate the PMP22 enhancer.
These assays demonstrated that wild-type EGR2 was able to increase expression of Fluc (~110 fold increase over the empty vector; Fig. 4) consistent with previous reports that the PMP22 enhancer includes an EGR2 response element. In contrast, p.Asp411Gly EGR2 induced Fluc activity at a significantly lower level (~40 fold increase over the empty vector p = 2.1 × 10−13 compared to wild-type; Fig. 4) indicating that the p.Asp411Gly variant reduces the capacity for EGR2 to appropriately activate the PMP22 enhancer. These results reveal a functional effect of p.Asp411Gly EGR2 and support the pathogenicity of this variant.
The p.Asp411Gly EGR2 variant decreases transcriptional activation. Wild-type and p.Asp411Gly EGR2 were evaluated for regulatory activity on a previously reported EGR2 response element at the PMP22 locus. Constructs (conditions indicated along the bottom) were transfected into cultured Schwann (S16) cells and tested for activity in luciferase assays normalized to an empty vector containing no genomic insert (far left column). Wild-type EGR2 and p.Asp411Gly EGR2 were each tested using two independently generated expression constructs (A and B) and each condition was tested in at least 24 biological replicates. The fold induction of Fluc activity is indicated along the y-axis and error bars indicate standard deviations. Statistical significance (p-value) was assessed using a two-tailed Student’s T-test.
Discussion
The spectrum of peripheral neuropathy associated with disease-causing EGR2 variants ranges from severe early onset DSN and CHN, to later onset demyelinating CMT1 and axonal CMT2. Using WES and functional testing we have determined that a de novo missense EGR2 variant, c.1232A > G p.Asp411Gly, causes a severe and early onset DSN phenotype by reducing the capacity for EGR2 to function as a transcription factor. This variant can be classified as ‘pathogenic’ according to guidelines determined by the American College of Medical Genetics and Genomics (PS2, PS3, PM1, PM2, PP2, PP3)33. This pathogenic variant affects the third zinc-finger domain of EGR2, which is highly conserved among species and is a DNA-binding motif crucial for its role as a transcription factor in the regulation of myelin genes34. All reported pathogenic variants are within these three zinc-finger regions except for p.Ile268Asn, which causes recessive CHN and is within an inhibitory domain (Table 3)4.
Functional testing conducted here with luciferase-based reporter assays demonstrated that p.Asp411Gly EGR2 activates transcription less effectively than wild-type EGR2, similar to what has been previously reported for pathogenic EGR2 variants located in the zinc-finger domains13,15,34,35. EGR2 function is crucial for myelination of the peripheral nervous system, and Krox20−/− (murine EGR2 orthologue) mice demonstrated arrested differentiation of Schwann cells at an early stage17. Interestingly, heterozygous Krox20+/− mice are phenotypically normal17, yet all reported pathogenic EGR2 variants in the zinc-finger domains cause peripheral neuropathy in a heterozygous state (Table 3)13. This is likely explained by the finding that in addition to pathogenic EGR2 variants resulting in a decrease or loss of EGR2 function, it has been shown that pathogenic EGR2 variants in all three zinc-finger domains (p.Arg359Trp, and p.Ser382Arg + p.Asp383Tyr, and p.Arg409Trp) result in the dominant-negative inhibition of wild-type EGR215,18 and SOX1018. These results are consistent with the reduction in transcriptional activation we observed in our luciferase assays with p.Asp411Gly EGR2.
The severity and onset of peripheral neuropathy caused by EGR2 variants is highly heterogenous, and it is yet to be determined why this broad phenotypic variability exists. This variation is exemplified by the p.Arg359Trp variant, which causes demyelinating CMT1D3, DSN5,36, and CHN5. This is also true for different variants of the amino acid adjacent to the p.Asp411Gly variant we have reported here, where p.Glu412Gly causes both demyelinating CMT1D12 and axonal CMT14, and p.Glu412Lys causes DSN35. Given these observations, it is possible that additional genetic factors exist which modify the phenotype of EGR2 variant, such as those recently reported for CMT1A37.
Our patient had findings on brain MRI and evoked potentials that may be consistent with multiple sclerosis, however it was determined that he did not fulfil diagnostic criteria. White matter lesions have been reported in some patients with CMT38,39,40,41, and the proband had no contrast enhancing lesions suggesting active demyelinating disease in both brain MRIs conducted. In CMTX1, which is caused by pathogenic variants in GJB1, white matter lesions were reported in 7.5% of a large cohort42. Given that EGR2 mediates transcription of GJB1, it is possible that the reduced efficiency of p.Asp411Gly EGR2 in activating transcription resulting in a phenotype similar to that seen in some CMTX1 patients. Additionally, the FMH phenotype may be a chance association, or due to another genetic cause that has not been identified, as there were no variants in hemiplegic migraine genes that segregated with the phenotype. White matter lesions have also been reported in patients with migraines and patients with sporadic hemiplegic migraine43,44,45,46, further complicating the phenotype seen in our proband.
In this study we report a novel de novo EGR2 variant, c.1232A > G p.Asp411Gly, as the cause of severe, early-onset CMT3 (DSN). We provide robust functional data to support the deleterious functional effect of this mutation. The phenotype is complicated by clinical features of white matter lesions and FMH which may be a chance association, although it would be important to consider the possibility of central nervous system involvement in future cases of EGR2-related neuropathies.
Methods
Subjects
The proband and three family members were recruited and informed consent was obtained for this study using protocols approved by the Sydney Local Health District Human Ethics Research Committee (SLHD HERC). All experiments were performed in accordance with relevant guidelines and regulations from SLHD HREC. Genomic DNA was extracted from peripheral blood using the PureGene Kit (Qiagen) following manufacturer’s instructions.
Whole-exome sequencing (WES)
WES was performed on genomic DNA (2.5 µg) in the proband and was outsourced to Macrogen (South Korea) as previously described19.
Bioinformatic analysis
Evolutionary conservation of the EGR2 D11 amino acid reside was conducted using GERP (http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html), phastCons and PhyloP (http://compgen.cshl.edu/phastweb/runtool.php). The predicted functional effect of the EGR2 p.Asp411Gly mutation was assessed using Polyphen2 (http://genetics.bwh.harvard.edu/pph2/), PROVEAN (http://provean.jcvi.org/seq_submit.php), SIFT (https://sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html), MutationAssessor (http://mutationassessor.org/r3/), and MutationTaster2 (http://www.mutationtaster.org/).
Sanger sequencing
All primers and PCR conditions are available upon request. PCR amplicons were sent to Garvan Molecular Genetics, Garvan Institute (Sydney, Australia) for Sanger sequencing using BigDye Terminator cycle sequencing protocols and visualised using Sequencher 2.3 software (Gene Codes Corporation).
Haplotype analysis
PCR amplicons from microsatellite markers (D1S347, D1S249, D2S2228, D2S140, D16S519) were sent for fragment analysis at Garvan Molecular Genetics, Garvan Institute (Sydney, Australia), and visualised using GeneMarker (SoftGenetics). Microsatellite marker size was determined using GS600 size standard.
Luciferase reporter gene expression constructs
Mutagenic primers were designed to model the p.Asp411Gly EGR2 mutation in an expression construct containing the mouse EGR2 open-reading frame directed by the CMV promoter (gift of John Svaren, University of Wisconsin). Site-directed mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and the manufacturer’s instructions. Mutant constructs were verified by Sanger sequencing (University of Michigan Medical School DNA Sequencing Core).
Cell culture, transfection, and luciferase assays
Cultured rat Schwann cells (S16)47 were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% (v/v) fetal bovine serum (Gibco), 2 mM L-glutamine (Gibco), 50 U/mL penicillin, and 50 g/mL streptomycin. For luciferase assays, ~1 × 104 S16 cells were plated in each well of a 96-well untreated cell culture plate and grown overnight at 37 °C with 5% CO2. After 24 h, the cells were transfected with the experimental constructs using Lipofectamine 2000 (Invitrogen) diluted 1:100 in OptiMem (Life Technologies). Cells in each well received 200 ng of an expression construct: either empty pEIB (expression vector containing a Fluc reporter gene with no genomic insert)32 or the pEIB-Forward vector with a PMP22 enhancer31 upstream of a Fluc reporter gene. S16 cells were co-transfected with 100 ng per well of wild-type or mutant EGR2 constructs in addition to the PMP22 enhancer construct. For an internal control for transfection efficiency, 2 ng of a pCMV-Rluc construct were transfected into cells in all wells. Each experimental construct was diluted in OptiMem and incubated with an equal volume of Lipofectamine 2000 in OptiMem for 20 min at room temperature before being applied to cells. Cells were incubated with transfection reagents for 4 h at 37 °C with 5% CO2, at which point the transfection reagents were removed and replaced with standard growth medium (above) and allowed to grow for 48 h at 37 °C with 5% CO2.
After 48 h, growth medium was removed, and cells were washed with 1X PBS. The cells were then lysed for 1 h at room temperature in 1X Passive Lysis Buffer (Promega), and 10 uL of lysate from each well was transferred into a white polystyrene 96-well plate (Corning). A Dual Luciferase Assay was performed using a Glomax Multi-Detection System and a Dual Luciferase Reporter 1000 Assay System kit (Promega) to determine Fluc and Rluc activities.
The ratio of Fluc to RLuc activity in each well was calculated, and ratios from individual wells were normalized relative to the average Fluc to Rluc ratio from the empty pEIB vector. The mean normalized ratio is a readout for the fold induction of Fluc by each experimental construct, and this is shown with standard deviation in the figure. Each variant was tested with two independently generated constructs (A and B in the figure) to rule out expression changes due to variation in the expression construct backbone, and each construct was tested in at least 24 individual experiments. Statistical significance was assessed using a two-tailed Student’s T-test.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Timmerman, V. et al. Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology 52, 1827–32 (1999).
Numakura, C. et al. Screening of the early growth response 2 gene in Japanese patients with Charcot-Marie-Tooth disease type 1. J. Neurol. Sci. 210, 61–4 (2003).
Chung, K. W. et al. Two missense mutations of EGR2 R359W and GJB1 V136A in a Charcot-Marie-Tooth disease family. Neurogenetics 6, 159–63 (2005).
Warner, L. E. et al. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 18, 382–384 (1998).
Boerkoel, C. F., Takashima, H., Bacino, C. A., Daentl, D. & Lupski, J. R. EGR2 mutation R359W causes a spectrum of Dejerine-Sottas neuropathy. Neurogenetics 3, 153–7 (2001).
Bellone, E. et al. A novel mutation (D305V) in the early growth response 2 gene is associated with severe Charcot-Marie-Tooth type 1 disease. Hum. Mutat. 14, 353–354 (1999).
Pareyson, D. et al. Cranial nerve involvement in CMT disease type 1 due to early growth response 2 gene mutation. Neurology 54, 1696–8 (2000).
Yoshihara, T. et al. A novel missense mutation in the early growth response 2 gene associated with late-onset Charcot–Marie–Tooth disease type 1. J. Neurol. Sci. 184, 149–153 (2001).
Mikešová, E. et al. Novel EGR2 mutation R359Q is associated with CMT type 1 and progressive scoliosis. Neuromuscul. Disord. 15, 764–767 (2005).
Shiga, K. et al. A novel EGR2 mutation within a family with a mild demyelinating form of Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst. 17, 206–209 (2012).
Briani, C., Taioli, F., Lucchetta, M., Bombardi, R. & Fabrizi, G. M. Adult onset Charcot-Marie-Tooth disease type 1D with an Arg381Cys mutation of EGR2. Muscle Nerve 41, 888–9 (2010).
Šafka Brožková, D., Nevšímalová, S., Mazanec, R., Rautenstrauss, B. & Seeman, P. Charcot–Marie–Tooth neuropathy due to a novel EGR2 gene mutation with mild phenotype – Usefulness of human mapping chip linkage analysis in a Czech family. Neuromuscul. Disord. 22, 742–746 (2012).
Sevilla, T. et al. The EGR2 gene is involved in axonal Charcot−Marie−Tooth disease. Eur. J. Neurol. 22, 1548–1555 (2015).
Tozza, S. et al. A novel family with axonal Charcot-Marie-Tooth disease caused by a mutation in the EGR2 gene. J. Peripher. Nerv. Syst. 24, 219–223 (2019).
Nagarajan, R. et al. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron 30, 355–68 (2001).
Jang, S.-W. et al. Locus-wide identification of Egr2/Krox20 regulatory targets in myelin genes. J. Neurochem. 115, 1409–20 (2010).
Topilko, P. et al. Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 (1994).
LeBlanc, S. E., Ward, R. M. & Svaren, J. Neuropathy-Associated Egr2 Mutants Disrupt Cooperative Activation of Myelin Protein Zero by Egr2 and Sox10. Mol. Cell. Biol. 27, 3521–3529 (2007).
Drew, A. P. et al. Improved inherited peripheral neuropathy genetic diagnosis by whole-exome sequencing. Mol. Genet. Genomic Med. 3, 143–154 (2015).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–11 (2001).
Gibbs, R. A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Karczewski, K. J. et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 531210; https://doi.org/10.1101/531210 (2019).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6, e1001025 (2010).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–50 (2005).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS One 7, e46688 (2012).
Sim, N. L. et al. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–7 (2012).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362 (2014).
Jones, E. A. et al. Distal enhancers upstream of the Charcot-Marie-Tooth type 1A disease gene PMP22. Hum. Mol. Genet. 21, 1581–1591 (2012).
Antonellis, A. et al. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 15, 259–71 (2006).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Warner, L. E., Svaren, J., Milbrandt, J. & Lupski, J. R. Functional consequences of mutations in the early growth response 2 gene (EGR2) correlate with severity of human myelinopathies. Hum. Mol. Genet. 8, 1245–51 (1999).
Szigeti, K. et al. Functional, histopathologic and natural history study of neuropathy associated with EGR2 mutations. Neurogenetics 8, 257–262 (2007).
Gargaun, E. et al. EGR2 mutation enhances phenotype spectrum of Dejerine–Sottas syndrome. Journal of Neurology 263, 1456–1458 (2016).
Tao, F. et al. Variation in SIPA1L2 is correlated with phenotype modification in Charcot- Marie- Tooth disease type 1A. Ann. Neurol. 85, 316–330 (2019).
Brockmann, K. et al. Cerebral involvement in axonal Charcot-Marie-Tooth neuropathy caused by mitofusin2 mutations. J. Neurol. 255, 1049–1058 (2008).
Lee, M.-J. et al. Six novel connexin32 (GJB1) mutations in X-linked Charcot-Marie-Tooth disease. J. Neurol. Neurosurg. Psychiatry 73, 304–6 (2002).
Luo, S., Jin, H., Chen, J. & Zhang, L. A Novel Variant in Non-coding Region of GJB1 Is Associated With X-Linked Charcot-Marie-Tooth Disease Type 1 and Transient CNS Symptoms. Front. Neurol. 10, 413 (2019).
Sacco, S., Totaro, R., Bastianello, S., Marini, C. & Carolei, A. Brain White Matter Lesions in an Italian Family with Charcot-Marie-Tooth Disease. Eur. Neurol. 51, 168–171 (2004).
Vivekanandam, V., Hoskote, C., Rossor, A. M. & Reilly, M. M. CNS phenotype in X linked Charcot- Marie-Tooth disease. J. Neurol. Neurosurg. Psychiatry 90, 1068 (2019).
Bashir, A., Lipton, R. B., Ashina, S. & Ashina, M. Migraine and structural changes in the brain. Neurology 81, 1260–1268 (2013).
Nagarajan, E., Bollu, P. C., Manjamalai, S., Yelam, A. & Qureshi, A. I. White Matter Hyperintensities in Patients with Sporadic Hemiplegic Migraine. J. Neuroimaging 29, 730–736 (2019).
Swartz, R. H. & Kern, R. Z. Migraine is associated with magnetic resonance imaging white matter abnormalities: A meta-analysis. Archives of Neurology 61, 1366–1368 (2004).
Kruit, M. C. et al. Migraine as a Risk Factor for Subclinical Brain Lesions. J. Am. Med. Assoc. 291, 427–434 (2004).
Toda, K., Small, J. A., Goda, S. & Quarles, R. H. Biochemical and cellular properties of three immortalized Schwann cell lines expressing different levels of the myelin-associated glycoprotein. J. Neurochem. 63, 1646–57 (1994).
Nakamura, T. et al. Vincristine exacerbates asymptomatic Charcot-Marie-Tooth disease with a novel EGR2 mutation. Neurogenetics 13, 77–82 (2012).
Wang, W. et al. Target-enrichment sequencing and copy number evaluation in inherited polyneuropathy. Neurology 86, 1762–1771 (2016).
Fusco, C. et al. Charcot-Marie-Tooth disease with pyramidal features due to a new mutation of EGR2 gene. Acta Biomed. 90, 104–107 (2019).
Acknowledgements
This work was supported by the Australian National Health and Medical Research Council project grant (APP1046690) awarded to M.L.K. and G.A.N. A.A. was supported by the National Institute of Neurological Disorders and Stroke (NS073748). We wish to acknowledge A/Prof. Alastair Corbett for conducting the nerve conduction study. We thank Prof John Svaren (University of Wisconsin) for the wild-type EGR2 expression construct. We thank the family members participating in this study.
Author information
Authors and Affiliations
Contributions
B.G., N.G. and M.E. carried out the experiments. G.N. and K.K. collected patient data. A.A. and M.K. supervised the project. M.K. conceived the original idea. B.G. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grosz, B.R., Golovchenko, N.B., Ellis, M. et al. A de novo EGR2 variant, c.1232A > G p.Asp411Gly, causes severe early-onset Charcot-Marie-Tooth Neuropathy Type 3 (Dejerine-Sottas Neuropathy). Sci Rep 9, 19336 (2019). https://doi.org/10.1038/s41598-019-55875-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-55875-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.