Abstract
In this study, we report the molecular structure of the breakpoint region in a new chromosomal translocation, t(2;7)(p24.3;p14.2), in a case of acute myeloid leukemia transformed from myelodysplastic syndrome (MDS). An extensive fluorescence in situ hybridization (FISH) analysis showed that NAG (2p24.3) and ELMO1 (7p14.2) were involved at the breakpoints of t(2;7)(p24.3;p14.2). Furthermore, we detected a novel chimeric transcript consisting of NAG and ELMO1. Interestingly, this transcript encoded a truncated molecular form of 3′ELMO1 as the result of a frameshift caused by the chromosomal translocation. Although this study does not provide direct evidence that a defect in NAG-ELMO1 plays a role in the pathogenesis or the leukemic change in MDS, it does suggest that defects in NAG-ELMO1 potentially contributed to the leukemic progression in this case.
Similar content being viewed by others
Introduction
Myelodysplastic syndrome (MDS) is a clonal hematopoietic stem-cell disorder characterized by ineffective hematopoiesis, trilineage dysplasia, marrow dysfunction and susceptibility to acute myeloid leukemia (AML).1, 2, 3, 4, 5, 6 Although the molecular mechanisms of MDS and its progression to AML have not been clarified, frequent chromosomal abnormalities, such as an entire or a partial deletion of the long arm of chromosome 5 or 7 (−5/5q− or −7/7q−), trisomy 8, der(1;7), deletion 20 and Y-missing, have been reported, suggesting an unidentified oncogene and/or tumor-suppressor gene to be involved in the pathogenesis and/or progression of this disease.5, 7 Balanced-type chromosomal translocations are infrequently observed in MDS, for example, t(5;12)(q33;p13), t(11;16)(q23;p13) and t(3;21)(q26;q21),6, 8 generating TEL/PDGFR-β,9 MLL/CBP10 and EVI-1/AML111 chimeric genes, respectively, which are involved in this disorder. Specific chromosomal abnormalities, especially translocations, found in hematological and nonhematological malignancies have provided important insights into the pathogenesis of malignant transformation, prompting us to explore and characterize novel chromosomal translocations in MDS or MDS-derived AML to understand the pathogenesis as well as to establish novel therapeutic strategies of this malignancy.
Here, we report a new chromosomal translocation, t(2;7)(p24.3;p14.2), observed in leukemic cells in a patient with AML transformed from MDS. Through a detailed characterization of the molecular structure of the break points involved in this translocation, we have identified a novel chimeric transcript consisting of the neuroblastoma-amplified gene (NAG)12, 13 at 2p24.3 and the engulfment cell motility gene 1 (ELMO1)14 at 7p14.2. The expression of NAG-ELMO1, in which the 3′ terminal of ELMO1 was truncated as a result of a frameshift caused by the chromosomal translocation, was detected.
Materials and methods
Case report and chromosomal analysis
A 62-year-old man was admitted to hospital in May 1997 with fever and pneumonia. At diagnosis, his white blood cell count was 1800 μl−1, hemoglobin level was 4.6 g dl−1 and platelet count was 30 000 μl−1, indicating pancytopenia. Bone marrow aspirated with informed consent was hypocellular with 17.6% blasts, expressing CD13, CD33 and CD34. On the basis of these findings and a normal cytogenetic pattern, he was diagnosed as having myelodysplastic syndrome (RAEB) with a normal karyotype, 46,XY and treated with granulocyte colony-stimulating factor. After 5 months, he developed AML-M4 with t(2;7)(p24.3;p14.2) in all 20 bone marrow metaphase cells that were analyzed (Figure 1), and received reinduction therapy with daunorubicin, cytarabine, 6-mercaptopurine and prednisolone. Although he temporarily achieved complete remission, he died 52 months after diagnosis from hypocellular marrow with atypical myeloid cells and serious pancytopenia. The translocation t(2;7)(p24.3;p14.2) was found as the sole anomaly, which disappeared during remission and reappeared at a relapse of the disease.
Fluorescence in situ hybridization (FISH)
FISH was carried out as described elsewhere.15 According to information from human genome databases (http://www.ncbi.nlm.nih.gov/ and http://genome.ucsc.edu/), we selected and used a total of 12 and 17 bacterial artificial chromosome clones (BACs) and P1-derived artificial chromosome clones (PACs) as probes, respectively, spanning the breakpoints of t(2;7)(p24.3;p14.2), and made contig maps around the break points (Figure 2).
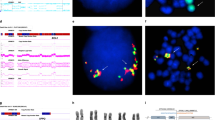
Maps around break points and representative results of metaphase FISH experiments. (a and d) The positions, according to the human genome database (http://genome.ucsc.edu/), of bases, genes and the BACs/PACs used as probes for FISH are represented to the right of chromosomes 7 (a) and 2 (d) ideograms. Open boxes, black boxes and oblique-lined boxes represent clones located on the partner chromosomes, clones remaining on the original chromosomes and clones with an entirely or partially deleted signal in FISH, respectively. The checked box represents the clone with a split signal pattern on FISH. A closed arrow indicates the possibly deleted region on chromosome 7. (b) Representative result of two-color FISH using BAC RP11-302L6 (green) and BAC RP11-164E6 (red). Green signals were observed on normal chromosome 7 and der(7), whereas red was observed only on normal chromosome 7. (c) Representative result of FISH using BAC RP11-59F6 (red). Red signals were weakly observed on the der(7) chromosome compared with normal chromosome 7. (e) Representative result of FISH using BAC RP11-516B14 (red). Red signals were observed on normal chromosome 2 as well as der(7) and der(2) (upper panel). Enlarged images of normal chromosomes 2 and 7 and those of der(2) and der(7) are shown in the lower panel.
Reverse-transcription (RT)-PCR and direct sequence analysis
Mononuclear cells were separated from BM samples with Lymphoprep (Nycomed pharma, Oslo, Norway). Total RNA was extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and single-stranded cDNA was synthesized using SuperScript First Strand Synthesis (Invitrogen). RT-PCR was carried out using FastStart Taq (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's instructions, using a series of primer sets designed on the basis of the sequences of BAC/PACs (http://www.ncbi.nlm.nih.gov/). All primer sequences are available on request. Each PCR product was separated by gel electrophoresis. Direct sequencing of the RT-PCR product was carried out with a Cyclist DNA sequencing kit (Stratagene, La Jolla, CA, USA). The results of the sequencing analysis were compared using the Basic Local Alignment Search Tool (BLAST) of the NCBI (http://www.ncbi.nlm.nih.gov/).
Results and discussion
The karyotype of mononuclear cells from the bone marrow aspirate of this patient at AML transformation showed a reciprocal translocation, possessing a break point on the short arm of chromosome 2 and the short arm of chromosome 7. As we first presumed his t(2;7) from a variant of t(7;11)(p15;p15), which is known to generate the HOXA9/NAP98 fusion gene in myeloid leukemia,16, 17 we conducted a FISH analysis using the PAC clone, RP1-170O19, containing the HOXA9 gene. However, clear signals of this clone were located on normal chromosome 7 and der(2)t(2;7), indicating that the break point of der(7) lies proximal to HOXA9. Thus, we carried out a series of FISH analyses using additional 9 BAC clones. Signals from BAC RP11-7P19 to BAC RP11-340F1 were translocated to der(2) (Figure 2a and data not shown), whereas the signal from BAC RP11-164E6 was detected on normal chromosome 7 but not on der(2) or der(7) t(2;7) (Figure 2b). Interestingly, the signal from BAC RP11-59F6 that was observed on der(7) was weaker than that on normal chromosome 7 (Figure 2c), suggesting a cryptic deletion to be involved. According to the human genome database (http://www.ncbi.nlm.nih.gov/ and http://genome.ucsc.edu/), only ELMO1, a human ortholog of the Caenorhabditis elegans gene, Ced12,14 is located around the break point of der(7) (Figure 2a), suggesting ELMO1 to be involved in the rearrangement and/or deletion at break point.
Next, we carried out FISH to determine the break point involved in der(2)t(2;7)(p24.3;p14.2). As shown in Figure 2d, the clones between BAC RP11-375P12 and BAC RP11-314E10 were translocated to der(7), whereas the clones from BAC RP11-65H13 to BAC RP11-111P18 were retained on der(2). Notably, BAC RP11-516B4 showed signals split between der(2) and der(7) (Figure 2e). According to the human genome database (http://www.ncbi.nlm.nih.gov/ and http://genome.ucsc.edu/), only NAG is located within this clone (Figure 2d), suggesting NAG to be involved in the break point of der(2)t(2;7)(p24.3;p14.2) in this case.
As (a) ELMO1 or NAG is located around each of the break points involved in this t(2;7)(p24.3;p14.2) and (b) these two genes are arranged in the same orientation, we speculated that a rearrangement of ELMO1 and NAG might generate a chimeric gene through the chromosomal translocation. To test this hypothesis, we tried to connect exons by RT-PCR using a series of primer sets designed on the basis of the 5′ of NAG and 3′ of ELMO1 or the 5′ of ELMO1 and 3′ of NAG. An RT-PCR product of minimal size was detected using the former set between exon 38 of NAG and exon 14 of ELMO1, but not the latter set between exon 13 of ELMO1 and exon 39 of NAG (Figure 3a). Sequencing of the product showed that the 3′ end of exon 38 of NAG was fused to the 5′ end of exon 14 of ELMO1 in the chimeric transcript, resulting in a truncated deduced amino acid sequence of the 3′-portion of ELMO1 because of a frameshift of the coding sequence of this gene (Figure 3b). The expression of this chimeric transcript was confirmed by RT-PCR, using primer sets designed to connect the possible start codon of NAG and possible novel stop codon of ELMO1 and sequencing of the RT-PCR product (Figure 3c), suggesting that a truncated fusion protein may be expressed in the leukemia cells of the patient. The deduced amino acid sequence of NAG-ELMO1 contains most of NAG and a very short additional sequence at the C terminus. As no functional characterization of the NAG protein has ever been reported and no functional motif has been mapped in this protein sequence, the functional significance of the truncated NAG-ELMO1 protein to leukemogenesis remains unclear. Our results of FISH analysis showed a cryptic deletion of the 5′-portion of ELMO1 through the translocation (Figures 2a–c), suggesting that the reciprocal ELMO1-NAG transcript was not expressed, possibly because of deletion of the 5′-portion of ELMO1. The expression of intact NAG and ELMO1, which are possibly transcribed from intact chromosomes 2 and 7, respectively, was detected by RT-PCR (data not shown).
Structure of the fusion point of the t(2;7)(p24.3;p14.2) translocation. (a) Identification of the NAG-ELMO1 chimeric transcripts by RT-PCR in the patient. 1 Kb DNA ladder (Invitrogen) was used as a size marker. The NAG-ELMO1 transcript was detected by RT-PCR, but the reciprocal ELMO1-NAG transcript was not. (b) Schematic representation of the NAG-ELMO1 mRNA junction and nucleotide and deduced amino acid sequences around the junction between exon 39 of NAG and exon 14 of ELMO1. Exon numbers are given above the map. Dotted boxes and gray boxes indicate exons of NAG and ELMO1, respectively. The positions and orientations of primers used in (a) are indicated by arrowheads. The NAG-ELMO1 transcript generated by t(2;7)(p24.3;p14.2) was a truncated molecular form of 3′ ELMO1, produced as the result of a frameshift. (c) Deduced amino acid sequence of the NAG-ELMO1 chimeric transcript. An open box indicates the sequence from ELMO1. The asterisk indicates the TGA termination codon.
MDS is a heterogeneous disorder, with several specific chromosomal abnormalities involved in its pathogenesis. However, it is relatively rare for a chromosomal translocation to affect the pathogenesis of AML developed from MDS. Recently, a monocytic leukemia cell line, ELAM1, was established from a patient with AML that had progressed from MDS.18 Cytogenetically, both the leukemic cells of the patient and ELAM1 cells indicated t(11;21)(q13;q22). The fusion transcripts of RUNX1/LRP16 and the reciprocal LRP16/RUNX1 were also detected in ELAM1 cells. Although the function of LRP16 remains unknown, the chromosomal translocation, t(11;21), is thought to contribute to the onset of the leukemic transformation from MDS, as this translocation is reported to disappear during remission and to reappear at relapse. In relation to the leukemic transformation of fusion genes, animal models have provided important insights into the molecular mechanisms underlying the progression from MDS to AML. Using a NUP98-HOXD13 fusion gene, Lin et al.19 developed a mouse model with all the key clinical features of MDS, including peripheral blood cytopenias, bone marrow dysplasia and apoptosis and transformation to AML. In contrast, the AML1/RUNX1 gene point mutation has been shown to be one of the major causes of refractory anemia with an excess of blasts (RAEB), RAEB in transformation (RAEB-t) and AML, following the molecular alteration of MDS.20 We did not investigate the AML1/RUNX1 gene point mutation in this case.
Rosenfeld and List21 have suggested a multi-stepped pathogenesis of MDS. In the first step, progenitor cell injury gives rise to genomic instability. This instability leads to stepwise gene mutations and/or chromosomal aberrations, the subsequent accumulation of which results in the diverse features of MDS. According to this hypothesis, the translocation t(2;7) during the second stage of pathogenesis is involved in the leukemic transformation from MDS. Interestingly, Paulsson et al.22 reported a 1.8-Mb hemizygous deletion at 7p14.1, which was identified in a case with MDS transforming to AML, using high resolution array-based comparative genomic hybridization, although the deleted region was more centrimeric to ELMO1 and did not include this gene. Conversely, it is possible that t(2;7) observed in leukemic cells in this case was one of the random chromosomal aberrations that were already present in a subset of RAEB cells through genomic instability. Moreover, an unknown genetic event responsible for progression to an overt leukemia occurred in one of the t(2;7) marker-positive cells, resulting in a clonal expansion.
Two molecular mechanisms resulting from chromosomal translocations are known to produce hematological malignancies.23 The first mechanism involves the formation of a chimeric transcript that has a dominant-negative effect or strong tyrosine kinase activity. The second mechanism involves the regulation of gene functions by a promoter located upstream of target genes. Although there is no direct evidence that a defect in NAG-ELMO1 plays a role in the pathogenesis or leukemic transformation of MDS, our observation suggests that NAG-ELMO1 contributes to disease progression because of the consistent appearance of t(2;7) in leukemic cells. Exploration of the cytogenetic and genetic alterations in MDS may increase our understanding of the molecular mechanisms underlying this disease and aid in the development of novel therapeutic strategies.
References
Raskind, W. H., Tirumali, N., Jacobson, R., Singer, J. & Fialkow, P. J. Evidence for a multistep pathogenesis of myelodysplastic syndrome. Blood 63, 1318–1323 (1984).
Janssen, J. W. G., Buschle, M., Layton, M., Drexler, H. G., Lyons, J. & Van Den Berghe, H. et al. Clonal analysis of myelodysplastic syndromes: evidence of multipotent stem cell origin. Blood 73, 248–254 (1989).
Fenaux, P. Myelodysplastic syndromes. Hematol. Cell Ther. 38, 363–380 (1996).
Van Lom, K., Hagemeijer, A., Vandekerkhove, F., Smit, E. M. E. & Löwenberg, B. Cytogenetic clonality analysis: typical patterns in myelodysplastic syndrome and acute myeloid leukemia. Br. J. Haematol. 93, 594–600 (1996).
Mhawech, P. & Saleem, A. Myelodysplastic syndrome: review of the cytogenetic and molecular data. Crit. Rev. Oncol. Hematol. 40, 229–238 (2001).
Hirai, H. Molecular mechanism of myelodysplastic syndrome. Jap. J. Clin. Oncol. 33, 153–160 (2003).
Fenaux, P. Chromosome and molecular abnormalities in myelodysplastic syndrome. Int. J. Hematol. 73, 429–437 (2001).
Chen, Z., Morgan, R., Baer, M. R., Ligorsky, R. & Sandberg, A. A. Translocation (3;21) characterizes crises in myeloid stem cell disorders. Cancer Genet. Cytognet. 57, 153–159 (1991).
Golub, T. R., Barker, G. F., Lovett, M. & Gilliland, D. G. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosome translocation. Cell 77, 307–316 (1994).
Taki, T., Sako, M., Tsuchida, M. & Hayashi, Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood 89, 3945–3950 (1997).
Mitani, K., Ogawa, S., Tanaka, T., Miyoshi, H., Kurokawa, M., Mano, H. et al. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 13, 504–510 (1994).
Wimmer, K., Zhu, X. X., Lamb, B. J., Kuick, R., Ambros, P. F., Kovar, H. et al. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene 18, 233–238 (1999).
Scott, D. K., Board, J. R., Lu, X., Pearson, A. D. J., Kenyon, R. M. & Lunec, J. The neuroblastoma amplified gene, NAG: genomic structure and characterization of the 7.3 kb transcript predominantly expressed in neuroblastoma. Gene 307, 1–11 (2003).
Gumienny, T. L., Brugnera, E., Tosello-Trampont, A. C., Kinchen, J. M., Haney, L. B., Nishiwaki, K. et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 (2001).
Inazawa, J., Ariyama, T. & Abe, T. Physical ordering of three polymorphic DNA markers spanning the regions containing a tumor suppressor gene of renal cell carcinoma by three-color fluorescent in situ hybridization. Jpn. J. Cancer Res. 83, 1248–1252 (1992).
Borrow, J., Shearman, A. M., Stanton, Jr. V. P., Becher, R., Collins, T., Williams, A. J. et al. The (7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NUP98 and class homeoprotein HOXA9. Nat. Genet. 12, 159–167 (1996).
Nakamura, T., Largaespada, D. A., Lee, M. P., Johnson, L. A., Ohyashiki, K., Toyama, K. et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12, 154–158 (1996).
Imagama, S., Abe, A., Suzuki, M., Hayakawa, F., Katsumi, A., Emi, N. et al. LRP16 is fused to RUNX1 in monocytic leukemia cell line with t(11;21)(q13;q22). Eur. J. Haematol. 79, 25–31 (2007).
Lin, Y. W., Slape, C., Zhang, Z. & Aplan, P. D. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 106, 287–295 (2005).
Harada, H. & Harada, Y. Point mutations in the AML1/RUNX1 gene associated with myelodysplastic syndrome. Crit. Rev. Eukaryot. Gene Expr. 15, 183–196 (2005).
Rosenfeld, C. & List, A. A hypothesis for the pathogenesis of myelodysplastic syndromes: implications for new therapies. Leukemia 14, 2–8 (2000).
Paulsson, K., Heidenblad, M., Strömbeck, B., Staaf, J., Jönsson, G., Borg, A. et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia 20, 840–846 (2006).
Gasparini, P., Sozzi, G. & Pierotti, M. A. The role of chromosomal alterations in human cancer development. J. Cell. Biochem. 102, 320–331 (2007).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Fujita, K., Sanada, M., Harada, H. et al. Molecular cloning of t(2;7)(p24.3;p14.2), a novel chromosomal translocation in myelodysplastic syndrome-derived acute myeloid leukemia. J Hum Genet 54, 355–359 (2009). https://doi.org/10.1038/jhg.2009.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.40
Keywords
This article is cited by
-
Role of ELMO1 in inflammation and cancer—clinical implications
Cellular Oncology (2022)
-
PAN3–PSMA2 fusion resulting from a novel t(7;13)(p14;q12) chromosome translocation in a myelodysplastic syndrome that evolved into acute myeloid leukemia
Experimental Hematology & Oncology (2018)
-
DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung
Oncogene (2010)