Abstract
Fluoroquinolones are antimicrobial agents that are widely used clinically, but the increasing resistance of Acinetobacter baumannii (A. baumannii) to these agents is a matter of concern. We investigated mutant prevention concentrations (MPCs) of three fluoroquinolones, levofloxacin (LVX), pazufloxacin (PAZ) and ciprofloxacin (CIP). We analyzed an A. baumannii standard strain (ATCC19606) for mutation prevention indices (MPIs), MPCs and mutant selection windows as well as MICs of CIP, PAZ and LVX and compared the derived values with 34 A. baumannii strains collected in hospitals. In addition, A. baumannii standard strain (ATCC19606) fluoroquinolone-resistant mutants were investigated for gyrA and parC gene mutations. MPCs of CIP, prevention antibiotics concentration and LVX for A. baumannii ATCC19606 were 12.8, 5.6 and 2.8 μg ml−1 and their MPIs were 16, 8 and 4, respectively. Clinically isolated A. baumannii strains had CIP, PAZ and LVX MPC value ranges of 1–8, 1–16 and 0.5–2 μg ml−1 and their MPIs were 8, 8 and 4 μg ml−1. Single gyrA mutations (Ser83-Leu83) occurred in 18 resistant strains (48.7%) and single parC mutations (Ala79-Asp79 or (Ser80-Leu80) occurred in 8 resistant strains (21.6%), whereas gyrA and parC double mutations occurred in 2 (5.4%) of the resistant strains. MPC and MPI values of LVX were lower than that of CIP and PAZ. Single gyrA and parC mutations accounted for the majority of mutations (n=24), whereas double mutations occurred only in two strains.
Similar content being viewed by others
Introduction
A. baumannii is a common Gram-negative bacterium responsible for hospital-acquired hospital (nosocomial) infections1, 2, 3 and in the past fluoroquinolones were prescribed as empiric therapy. However, A. baumannii resistance changes to fluoroquinolones has become increasingly serious in recent years. A. baumannii resistance to fluoroquinolones is caused by mutations in the quinolone resistance-determining regions (QRDR), changed outer membrane protein expression and drug efflux pump overexpression.4, 5, 6 Fluoroquinolone mainly acts on DNA gyrase and topoisomerase IV, which are varied by gene mutations in the QRDR. DNA gyrase is a tetramer consisting of two A subunits and two B subunits, which are encoded by gyrA and gyrB. Topoisomerase IV is also a tetramer consisting of two C and two E subunits, which are encoded by parC and parE genes. Mutations may occur in all four genes, but they are predominantly located in gyrA and parC.7, 8 QRDR mutations are considered to be the main cause of A. baumannii resistance to fluoroquinolones.
Previously, a theory of mutant prevention antibiotics concentrations has been proposed.9, 10 The theory assumes that in a bacterial population some resistant mutant subpopulations already exist before the antimicrobial treatment. These drug-resistant mutant subpopulations are enriched and amplified during therapy when antimicrobial concentrations fall within a specific range, which is called the mutant selection window (MSW). The lower MSW concentration boundary is approximated by the MIC and the upper MSW concentration boundary is the lowest concentration for inhibiting the growth of non-susceptible, first-step mutant strains and is termed the mutant prevention concentration (MPC).11 MPC is also an antibacterial threshold concentration that limits the selective amplification of mutant strains. Bacteria only can grow with two, or more than two, mutations when the concentration of antibacterial drugs is above the MPC. The ratio between the MPC and MIC is called the mutation prevention index (MPI), which represents the opening of the MSW. In principle, a narrow window would expose the mutant subpopulation to selective drug concentrations for shorter times. A dosage regimen based on traditional pharmacodynamic standards uses drug concentrations that fall into the MSW and selectively enrich drug-resistant mutants.12 The MSW theory provides new directions for the dosage regimen of antibiotics, because after studying the specific drug MPC and pharmacokinetic parameters of a particular antibiotic, applying concentrations higher than the MPC can impede acquisition of drug resistance. It has been proposed that for situations in which antimicrobial concentrations cannot be kept above the MSW, a combination therapy should be employed to restrict the enrichment of mutants.13
Levofloxacin (LVX), pazufloxacin (PAZ) and ciprofloxacin (CIP) are fluoroquinolones that are commonly used in hospitals, but their actual MPC against A. baumannii is not clear and the spread of fluoroquinolone-resistant A. baumannii strains to Asia has been noted.14 In the present study, we evaluated the MPCs of three antibiotics against A. baumannii isolates from patients in our hospital, in order to determine the fluoroquinolone susceptibilities of mutant subpopulations.
Materials and Methods
Bacterial strains
The A. baumannii standard strain (ATCC19606) was purchased from the National Institute for the Control of Pharmaceutical and Biological Products in China. We further tested 34 A. baumannii strains susceptible to CIP, which were isolated from clinical specimens (23 from the respiratory tract and 11 from surgical wounds). Only one isolate from each patient was used in further analyses. For quality controls, we used Escherichia coli ATCC27853 with expected MIC values of 0.004–0.016 mg l−1 (CIP), 0.008–0.06 mg l−1 (LVX) and 0.008–0.06 mg l−1 (PAZ).
Antibiotics and chemicals
CIP (84.9% content) and LVX (97.3% content) were purchased from the Institute for the Control of Pharmaceutical and Biological Products in China, whereas PAZ (99.3% content) was obtained from the Xinhua pharmaceutical factory in Shandong province. The antibiotics were dissolved in double distilled water and stored at −80 °C as 2560 μg ml−1 stock solutions. Culture medium Mueller-Hinton (MH) agar, MH broth and soybean trypsin agar were purchased from OXOID (Thermo Fisher Scientific Inc., Hampshire, UK). Goodview type I nucleic acid dye, PrimeSTAR HS DNA Polymerase, during therapy mixture (each 2.5 mm), 10 × PrimeSTAR Buffer (Mg2+plus), DNA Marker DL2000, MiniBEST Bacterial Genomic DNA Extraction Kit Ver. 2.0, Agarose Gel DNA Purification Kit Ver. 2.0 and PCR primer were purchased from Bao (DaLian, China).
According to previous reports,15, 16 the ATCC19606 strain, which grew stably, was inoculated on plates with different drug dilutions and the number of bacterial colonies counted after overnight culture. MICs were first measured by the agar plate double-dilution method and then the MIC90 (the lowest concentration at which 90% of bacteria had a MIC value below the determined concentration) was evaluated by linearly decreasing drug concentrations by 10% based on the MIC values. The A. baumannii (>1010 CFU ml−1) standard strain was inoculated on serial agar plates containing different antibacterial concentrations above MIC. Specimens were incubated at 35 °C and the growth state observed at 24 , 48 and 72 h. The lowest antibacterial drug concentration at which no bacteria grew was determined and termed the provisional MPC (MPCpr). To correct the MPVpr for inoculum effects and to narrow the MPCs, the growing colonies were restreaked onto agar plates containing up to 20% of the MPCpr values of antibiotics concentrations, which were linearly decreased and increased as required. The lowest concentration at which no bacteria grew was taken to be the MPC.17
Measurements of MIC90 and MPC for fluoroquinolones against 34 strains of A. baumannii susceptible to CIP
Single bacterial colonies from plates after fresh overnight culture were adjusted to concentrations of 1.5 × 108 CFU ml−1 with physiological saline. Then, 200 μl of the solution was added to 1.8 ml of physiological saline and inoculated on plates, using a 1 μl quantitative inoculating loop, so that each plate would contain ~104 bacteria. The diluted test bacteria and quality control bacterial were all inoculated within 15 min and the plates were incubated for 18 h at 35 °C when MICs were determined. The lowest growth-inhibiting concentration was considered to be the MIC according to the USA Clinical and Laboratory Standards Institute standard.18 The measurements of MPCs were carried out as described above.
Selection of standard strain ATCC19606-resistant mutants and measurement of their MICs
A single bacterial colony from a plate containing the standard A. baumannii strain ATCC19606, after fresh overnight culture, was selected and inoculated in 20 ml MH broth for culture in a rotator overnight. Samples were then added to 200 ml MH broth and cultured for a further 6 h. After centrifugation at 3000 r.p.m. the bacterial solution was ~6–9 × 109 CFU ml−1, which was then adjusted to 3 × 1010 CFU ml−1. The solution was inoculated onto MH agar plates containing two MIC-MPC multiple concentrations of antibacterial drugs and incubated for 72 h at 35 °C. Then 1–6 bacteria colonies from each MH agar plate were inoculated onto plates containing the primary drug concentrations for mutant selection and incubated for 16–18 h at 35 °C. If the selected bacteria still grew on these plates, the strains were considered to be resistant mutants and the MIC was determined by the method described above.
Gene sequencing of ATCC19606-resistant mutant against fluoroquinolones
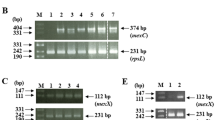
DNA of the above selected mutant was extracted following the instructions of the MiniBEST Bacterial Genomic DNA Extraction Kit Ver. 2.0 (Takara, Shiga, Japan). PCR primer sequences were designed as follows: the upstream primer of gyrA was 5′-AAATCTGCCCGTGTCGTTGGT-3′, the downstream primer of gyrA was 5′-GCCATACCTACGGCGATACC-3′ with an amplification length of 268 bp. The upstream primer of parC was 5′-CAGCGCCGTATTGTCTATGC-3′ and the downstream primer of parC was 5′-CCATTCGCTAGTGCCCTGAC-3′ with an amplification length of 263 bp. After PCR amplification, the resulting DNA fragments were separated by agarose gel electrophoresis, purified and stored at −20 °C. Finally, the purified DNA was sequenced and the data obtained before and after mutation were analyzed using GeneBank sequence alignment software (http://www.ncbi.nlm.nih.gov/BLAST/) to detect any mutations.
Results
MIC90, MPC and MSW of three fluoroquinolones against A. baumannii ATCC19606 standard strains
MICs of CIP, PAZ and LVX against A. baumannii ATCC19606 were all 1 μg ml−1, which was in an acceptable range for the quality control strain and MIC90 were 0.8, 0.7 and 0.7 μg ml−1, respectively. However, MPCs of CIP, PAZ and LVX for ATCC19606 were 12.8, 5.6 and 2.8 μg ml−1 and MPIs (MPC/MIC90) were 16, 8 and 4, respectively (Table 1).
Drug sensitivity determinations of 34 clinically isolated A. baumannii strains, which were susceptible to CIP, for three fluoroquinolones
No substantive differences of MIC ranges and MIC90 values of the three fluoroquinolones tested against the 34 strains of clinically isolated A. baumannii were detected. In contrast, MPCpr90 of CIP, PAZ and LVX were 4 , 8 and 1 μg ml−1 and MPI (MPCpr90/MIC90) values appeared as LVX<PAZ=CIP (Table 2). The data suggested that resistance mutations in clinically isolated A. baumannii strains might occur more easily with CIP and PAZ compared with LVX.
MIC measurement of A. baumannii ATCC19606 strain mutants
Thirty-seven mutant strains were selected for this study and MIC90 of the first-step mutant selections by CIP, PAZ and LVX were 8–16 μg ml−1 (4 x MIC, 8 x MIC), 2–8 μg ml−1 (2 x MIC, 4 x MIC) and 1–4 μg ml−1 (2 x MIC, 4 x MIC) (Table 3). MIC90 values of the mutants derived from the first-step selection against CIP and PAZ were high, suggesting that strong resistance has occurred, whereas the MIC90 values for LVX were low, suggesting they were still susceptible or only exhibited intermediate resistance to LVX.
DNA analyses of 37 A. baumannii ATCC19606 fluoroquinolone-resistant mutant strains
From 37 ATCC19606 A. baumannii-resistant mutant strains, we amplified 268 bp fragments of gyrA and 263 bp fragments of parC, both located in the QRDR regions, and compared the sequencing results with the respective wild-type sequences in GeneBank (accession number X82165 for gyrA and X95819 for parC). The results showed that 18 of the 37 (48.7%) first-step selection mutants had a Ser83 to Leu83 mutation as result of C-to-T substitutions of the nucleotide 246 in the gyrA gene. Mutations amplified by different selecting concentrations of LVX, CIP and PAZ were the same and 8 of the 37 (21.6%) first-step selection mutants had nucleotide substitutions at nucleotides 236 and 239 in the parC gene, resulting in six cases of Ala79 to Asp79 mutations and two cases of Ser80 to Leu80 mutations. In addition, 15 silent mutations occurred in the parC coding region (Table 4). Double mutations of gyrA and parC occurred in two mutant strains, accounting for 5.4% of the 37 first-step selection mutants. The MICs of the two mutant strains against CIP were 256 and 128 μg ml−1.
Taken together, 26 mutants showed changes in the DNA gyrase or topoisomerase IV gene and the remaining 11 strains presumably must have developed different resistance mutations.
Discussion
There is a paucity of information available about antibiotic MPCs for A. baumannii, and any published data are for carbapenems19 and colistin.20 In the present study, the MPCs of PAZ and CIP against the A. baumannii ATCC19606 strain were 2 and 4.6 × higher than that of LVX. The MPIs of PAZ and CIP were 2 and 4 × higher than that of LVX. These findings suggest that LVX is superior to PAZ and CIP in avoiding resistant mutant strain selection, data supported by the narrow MSW of LVX for the ATCC19606 strain compared with PAZ and CIP. Furthermore, the results from the ATCC19606 standard strains were consistent with those from the 34 CIP susceptible strains of A. baumannii. As the strains were clinically isolated, the results confirm the guiding value of a clinical drug regimen based on the ATCC19606 standard strain.
According to a previous report, area under the curve (AUC24)/MPC and Cmax/MPC values are important parameters for predicting the emergence of resistant mutants; the AUC24/MPC values should be >25 and Cmax/MPC ratios >2.2.21 AUC24 values for LVX have been reported to be 72.53 μg h−1ml−1 with Cmax of 8.67 μg ml−122 and 66.1 μg h−1ml−1 with a Cmax of 7.5 μg ml−1 23 after single 500 mg i.v. injections and 93 μg h−1 ml−1 with a Cmax of 11.3 μg ml−1 after a single 750 mg i.v. injection.24 Blood Cmax was 2.75μg ml−1 after a single i.v. administration of 200 mg LVX, with a t1/2 of 6.77 h in our measurements, which was in accordance with a previously reported concentration of 2.48 μg ml−1, a t1/2 of 6.95 h and an AUC value of 17.9 μg h −1ml−1 after a 200 mg LVX application.25 As the MPC for LVX was 2.8 μg ml−1 in our A. baumannii standard strain, the blood concentrations of LVX should be >6.16 μg ml−1 (2.2xMPC) the AUC >70 μg h−1ml−1 (25xMPC) and at least 500 mg LVX should be applied.
According to the FDA description of CIP, Cmax values are between 1.2 and 5.4 μg ml−1 after 250–1000 mg CIP applications and even a high dose of 1000 mg does not lead to a serum concentration above our calculated Cmax MPC of 12.8 μg ml−1 for the A. baumannii standard strain. These findings are supported by the fact that particular CIP resistance develops in A. baumannii during conventional CIP medication, but is unusual for Gram-negative bacteria against which CIP is generally more active than LVX.
Pazufloxacin Cmax serum concentrations were between 5.11 and 18.06 μg ml−1 with AUC values of 13.7–58.6 μg h−1 ml−1 after single 300–1000 mg i.v. pazufloxacin administrations.26 With our calculated MPC concentration of 5.6 μg ml−1 for the A. baumannii standard strain, the Cmax should be 12.32 μg ml−1 and the AUC24 140 μg h−1ml−1, which could be achieved by two applications of 1000 mg pazufloxacin per day (AUC 137 μg h−1 ml−1, Cmax 32 μg ml−1).27
Previous studies noted that QRDR gene mutations were the most important pathological mechanisms involved in microbe resistance to fluoroquinolones,28 which is also supported by 48.7% gyrA mutations and 21.6% parC mutations in the 37 first-step selected A. baumannii ATCC19606 mutants in the present study. Fluoroquinolone-resistance mutations were reported to be selected first in the more susceptible target genes, which is DNA gyrase in Gram-negative bacteria and topoisomerase IV in Gram-positive bacteria.29 The present data suggest, in contrast with other Gram-negative work, that the two enzyme targets have a similar affinity for the drugs. This assertion is likely to be true, particularly taking into account the multiple silent mutations in parC, which made the incidence of base mismatching during parC gene duplications even higher than that of the gyrA gene (18 versus 23).
However, gyrA and parC mutations did not occur in all mutant strains, which may be attributed to changes in outer membrane protein expression and drug efflux pumps. gyrA mutations mainly occurred between the N-terminal nucleotides 199 and 318 (encoding amino acids 67–106) and nucleotide changes causing Ser83 to hydrophobic Leu83 exchange. This change produces a reduced affinity for fluoroquinolones30 and comprised the majority of mutations in our study, which is in accordance with a previous report.31 In addition, other research groups have reported that gyrA mutations are the main factors causing resistance and that mutations in parC were additional factors for the development of A. baumannii resistance against CIP.32, 33
Saroj et al.34 found that expression of RecCBD in A. baumannii can increase, thereby strengthening the ability of repairing DNA damage caused by inhibition of DNA gyrase and topoisomerase IV, thereby increasing A. baumannii resistance to fluoroquinolones. Whether this mechanism is involved in high-level resistance to fluoroquinolones owing to single parC mutations should be analyzed further. However, Ala79-Asp (six strains) and Ser80-Leu (two strains) mutations in parC genes in the present study were slightly different from the main mutation site in Ser80 reported previously.35 Therefore, the role of Ala79-Asp mutations needs to be further clarified. Double mutations of gyrA and parC in two strains (the MICs of the two strains against CIP were 256 and 128 μg ml−1, respectively) resulted in high-level resistance, which was also noted in another study.36
We assessed fluoroquinolones for their property of selective amplification of A. baumannii-resistant mutant strains and indeed, high incidences of gyrA and parC mutations were detected in our study. Whether particular gyrA and parC mutations were the sole reason for A. baumannii resistance against fluoroquinolones needs to be examined further in future studies.
Conclusions
MPCs and MPIs can be used to assess effectively the concentration range in which fluoroquinolones selectively lead to A. baumannii-resistant mutant strain amplification. Of the drugs tested, LVX was superior to pazufloxacin and CIP as far as concentration regimens for mutant inhibition was concerned. The majority of LVX, pazufloxacin and CIP causing mutations manifested as Ser83 of the gyrA gene, but parC gene alterations also occurred, and double mutations in gyrA and parC genes resulted in the highest-level resistance against the fluoroquinolones.
References
Dijkshoorn,, L., Nemec,, A. & Seifert, H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951 (2007).
Gootz, T. D. & Marra, A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert. Rev. Anti. Infec. Ther. 6, 309–325 (2008).
Peleg, A. Y., Seifert, H. & Paterson, D. L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 (2008).
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P. & Perichon, B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393 (2010).
Lin, L., Ling, B. D. & Li, X. Z. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int. J. Antimicrob. Agents 33, 27–32 (2009).
Drlica, K. & Malik, M. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3, 249–282 (2003).
Casin, I., Breuil, J., Darchis, J. P., Guelpa, C. & Collatz, E. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg. Infect. Dis. 9, 1455–1457 (2003).
Randall, L. P., Coldham, N. G. & Woodward, M. J. Detection of mutations in Salmonella enterica gyrA, gyrB, parC and parE genes by denaturing high performance liquid chromatography (DHPLC) using standard HPLC instrumentation. J. Antimicrob. Chemother. 56, 619–623 (2005).
Zhao, X. & Drlica, K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin. Infect. Dis. 33, S147–S156 (2001).
Drlica, K. & Zhao, X. Mutant selection window hypothesis updated. Clin. Infect Dis. 44, 681–688 (2007).
Dong, Y., Zhao, X., Domagala, J. & Drlica, K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob. Agents Chemother. 43, 1756–1758 (1999).
Gebru, E., Damte, D., Choi, M. J., Lee, S. J., Kim, Y. H. & Park, S. C. Mutant prevention concentration and phenotypic and molecular basis of fluoroquinolone resistance in clinical isolates and in vitro-selected mutants of Escherichia coli from dogs. Vet. Microbiol 154, 384–394 (2012).
Drlica, K. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52, 11–17 (2003).
Yasuhara, T., Kugawa, S., Tateishi, Y., Ugajin, K., Yoshida, K. & Fukuchi, K. MLST analysis of multiple antimicrobial resistant Acinetobacter baumannii. Rinsho byori 61, 488–492 (2013).
Cui, J. C., Liu, Y. N. & Chen, L. A. Mutant prevention concentration of tigecycline for carbapenem-susceptible and -resistant Acinetobacter baumannii. J. Antibiot. (Tokyo) 63, 29–31 (2010).
Durante-Mangoni, E. & Zarrilli, R. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6, 407–422 (2011).
Drlica, K. A strategy for fighting antibiotic resistance. ASM News. 67, 27–33 (2001).
CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standards, 8th edn, CLSI document M07-A8 Clinical Laboratory Standards Institute: Wayne, PA, USA (2009).
Credito, K., Kosowska-Shick, K. & Appelbaum, P. C. Mutant prevention concentrations of four carbapenems against gram-negative rods. Antimicrob. Agents Chemother. 54, 2692–2695 (2010).
Cai, Y., Li, R., Liang, B., Bai, N., Liu, Y. & Wang, R. In vitro antimicrobial activity and mutant prevention concentration of colistin against Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 3998–3999 (2010).
Liang, B., Bai, N., Cai, Y., Wang, R., Drlica, K. & Zhao, X. Mutant prevention concentration-based pharmacokinetic/pharmacodynamic indices as dosing targets for suppressing the enrichment of levofloxacin-resistant subpopulations of Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 2409–2412 (2011).
Preston, S. L. et al. Levofloxacin population pharmacokinetics and creation of a demographic model for prediction of individual drug clearance in patients with serious community-acquired infection. Antimicrob. Agents Chemother. 42, 1098–1104 (1998).
Rebuck, J. A., Fish, D. N. & Abraham, E. Pharmacokinetics of intravenous and oral levofloxacin in critically ill adults in a medical intensive care unit. Pharmacotherapy 22, 1216–1225 (2002).
Kiser, T. H., Hoody, D. W., Obritsch, M. D., Wegzyn, C. O., Bauling, P. C. & Fish, D. N. Levofloxacin pharmacokinetics and pharmacodynamics in patients with severe burn injury. Antimicrob. Agents Chemother. 50, 1937–1945 (2006).
Lubasch, A., Keller, I., Borner, K., Koeppe, P. & Lode, H. Comparative pharmacokinetics of ciprofloxacin, gatifloxacin, grepafloxacin, levofloxacin, trovafloxacin, and moxifloxacin after single oral administration in healthy volunteers. Antimicrob. Agents Chemother. 44, 2600–2603 (2000).
Lee, J. et al. Single-dose pharmacokinetics and dose proportionality of intravenous pazufloxacin mesilate in healthy Korean volunteers. Expert Opin. Drug. Metab. Toxicol. 8, 921–928 (2012).
Kohno, S. et al. A clinical phase III study of pazufloxacin in patients with bacterial pneumonia. J. Jpn Soc. Chemother. 58, 664–680 (2010).
De la Fuente, C. M. et al. [Mutations in gyrA and gyrB genes among strains of Gram-negative bacilli isolated from Chilean hospitals and their relation with resistance to fluoroquinolones]. Rev. Med. Chil. 135, 1103–1110 (2007).
Jacoby, G. A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41, S120–S126 (2005).
Aldred, K. J., Kerns, R. J. & Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry. 53, 1565–1574 (2014).
Mak, J. K., Kim, M. J., Pham, J., Tapsall, J. & White, P. A. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 63, 47–54 (2009).
Hamouda, A. & Amyes, S. G. Novel gyrA and parC point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J. Antimicrob. Chemother. 54, 695–696 (2004).
Spence, R. P. & Towner, K. J. Frequencies and mechanisms of resistance to moxifloxacin in nosocomial isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 52, 687–690 (2003).
Saroj, S. D., Clemmer, K. M., Bonomo, R. A. & Rather, P. N. Novel mechanism for fluoroquinolone resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 4955–4957 (2012).
Vila, J., Ruiz, J., Goni, P. & Jimenez de Anta, T. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J. Antimicrob. Chemother. 39, 757–762 (1997).
Chenia, H. Y., Pillay, B. & Pillay, D. Analysis of the mechanisms of fluoroquinolone resistance in urinary tract pathogens. J. Antimicrob. Chemother. 58, 1274–1278 (2006).
Acknowledgements
We thank Professor Ye Mingliang, Dr Wang Xin and Li Jixia from the Jinan Region General Military Hospital for their critical evaluation of our manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sun, C., Hao, J., Dou, M. et al. Mutant prevention concentrations of levofloxacin, pazufloxacin and ciprofloxacin for A. baumannii and mutations in gyrA and parC genes. J Antibiot 68, 313–317 (2015). https://doi.org/10.1038/ja.2014.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.150