Abstract
Warming has been shown to cause soil carbon (C) loss in northern grasslands owing to accelerated microbial decomposition that offsets increased grass productivity. Yet, a multi-decadal survey indicated that the surface soil C stock in Tibetan alpine grasslands remained relatively stable. To investigate this inconsistency, we analyzed the feedback responses of soil microbial communities to simulated warming by soil transplant in Tibetan grasslands. Whereas microbial functional diversity decreased in response to warming, microbial community structure did not correlate with changes in temperature. The relative abundance of catabolic genes associated with nitrogen (N) and C cycling decreased with warming, most notably in genes encoding enzymes associated with more recalcitrant C substrates. By contrast, genes associated with C fixation increased in relative abundance. The relative abundance of genes associated with urease, glutamate dehydrogenase and ammonia monoxygenase (ureC, gdh and amoA) were significantly correlated with N2O efflux. These results suggest that unlike arid/semiarid grasslands, Tibetan grasslands maintain negative feedback mechanisms that preserve terrestrial C and N pools. To examine whether these trends were applicable to the whole plateau, we included these measurements in a model and verified that topsoil C stocks remained relatively stable. Thus, by establishing linkages between microbial metabolic potential and soil biogeochemical processes, we conclude that long-term C loss in Tibetan grasslands is ameliorated by a reduction in microbial decomposition of recalcitrant C substrates.
Similar content being viewed by others
Introduction
Global warming has a wide influence on terrestrial ecosystems, particularly in polar or alpine ecosystems, where it is predicted to be more pronounced (Walther et al., 2002). A number of field and modeling studies have indicated that climate warming causes soil carbon (C) loss in northern ecosystems (Melillo et al., 2002; Feng et al., 2008; Ise et al., 2008); yet, a plateau-wide survey of topsoil C stocks in Tibetan grasslands from 1980 to 2004 suggested that it was remarkably stable (Yang et al., 2009) in spite of climatic warming in this region, which is currently three times the global average (Li and Tang, 1988). This inconsistency suggests that biogeochemical processes must be stabilizing these C reservoirs; here we aim to elucidate the genetic mechanisms underpinning such activity. As microbes have an important role in mediating biogeochemical processes (Falkowski et al., 2008), we hypothesize that differential microbial functional capacity in C degradation and fixation might be responsible for this inconsistency.
The effects of climate warming can be simulated using artificial warming (Zhang et al., 2005), observational investigations along a temperature gradient (Dunne et al., 2004; Yang et al., 2014), and/or common garden transplant experiments (Waldrop and Firestone, 2006; Lazzaro et al., 2011; Zhao et al., 2014). Although artificial warming is straightforward and widely adopted, it can decrease soil moisture content, which conflicts with how we expect alpine and polar regions to respond to climate warming, as they will likely experience higher soil moisture owing to glacial and permafrost melt. Recently, soil transplants have been used to simulate the impact of climate change on terrestrial ecosystems (Balser and Firestone, 2005; Breeuwer et al., 2010; De Frenne et al., 2011; Lazzaro et al., 2011; Vanhala et al., 2011), with significant changes in microbial community structure observed with increased temperature (Vanhala et al., 2011). However, there are only limited studies, which provide evidence of the responses of microbial functional potential to soil warming.
Here we used common garden soil transplantation in alpine grassland on the Tibetan plateau, along an elevation gradient of 3200, 3400, 3600 and 3800 m above the sea level. Soil was transplanted from higher elevation to lower elevation plots to simulate warming, and microbial community structure and functions were profiled to determine what were the potential mechanisms underpinning C and nitrogen (N) metabolism, how microbially mediated environmental processes (for example, gas flux) correlated with changes in microbial gene abundance, and whether these changes could be modeled to extrapolate observed processes.
Materials and methods
The study site and experiment design
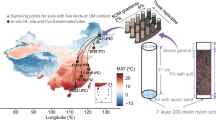
Four different elevations (3200, 3400, 3600 and 3800 m) along the southern slope of the Qilian Mountains, Qinghai, China, located in Haibei Alpine Meadow Ecosystem Research Station (37°37′N, 101°12′E) of the Northeastern Tibet Plateau, were used for the transplant experiment. The region is dominated by discontinuous permafrost (Zhuang et al., 2010), and the mean annual air temperature and rainfall precipitation is −1.7 °C and 560 mm, respectively (Zhao et al., 2006). The main soil type is Mat Cryic Cambisols, and the dominant aboveground vegetation is C3 herbal species. Soil pH at the depths of 10 and 20 cm is 7.3 and 7.4, respectively.
Experimental plots with sizes of 1 m length × 1 m width × 0.3 m depth were set up in May 2007. Along an elevation gradient of 3200, 3400, 3600 and 3800 m above the sea level, soil was transplanted in triplicates downward from higher to lower sites, resulting in a total of 18 transplanted plots (namely, 3800T3600, 3800T3400, 3800T3200, 3600T3400, 3600T3200 and 3400T3200). To minimize the disturbance to aboveground vegetation, four intact soils with the size of 0.5 m length × 0.5 m width × 0.3 m depth were dug out of the ground as a whole and pieced together to form a plot. Three plots at the elevations of 3400, 3600 and 3800 m were mock transplanted as they were dug out and then put back to the same places to serve as the control.
Soil samples were collected in August 2009 and used for GeoChip 4.0 experiments and environmental variable measurements. Three soil cores with a diameter of 1.5 cm at the depth of 0–20 cm were taken randomly at each plot. Then soil samples were transported to laboratory with ice and sieved with 2 mm mesh to remove visible grassroots and stones. Soil samples for GeoChip experiments were kept at −80 °C until DNA extraction, and soil samples for environmental variable measurements were kept at 4 °C or −20 °C.
Environmental variable measurements
Soil temperature was measured at the depths of 5, 10 and 20 cm using type-K thermocouples (Campbell Scientific, Logan, UT, USA) coupled to a CR1000 datalogger, whereas soil moisture was recorded every 30 min at the depths of 5 and 10 cm with time domain reflectometry (Model Diviner-2000, Sentek Pty Ltd, Stepney, SA, Australia). Soil biogeochemical variables were measured as previously described (Yang et al., 2014). In brief, total organic C (TOC) and total N (TN) were measured by a TOC-5000 A analyzer (Shimadzu Corp., Kyoto, Japan) and a Vario EL III Elemental Analyzer (Elementar, Hanau, Germany), respectively, as described previously (Ryba and Burgess, 2002, Stevens et al., 2012). NH4+-N, NO3−-N and greenhouse gas (CO2, N2O and CH4) effluxes were analyzed with a FIAstar 5000 Analyzer (FOSS, Hillerd, Danmark) and a gas chromatography (HP Series 4890D, Hewlett Packard, Wilmington, DE, USA), respectively. C/N ratio was calculated, dividing TOC by total N.
Soil bulk density at the depth of 0–20 cm was estimated by an empirical equation (1) using the corresponding TOC data at depths of 0–10 cm and 10–20 cm, whereas soil organic C density at the depth of 0–20 cm was calculated using equation (2), in which n was 2 for the two layers with soil thickness of 10 cm (Yang et al., 2009).


where SOCD, L, BD, TOC and M were soil organic C density (kg Cm−2), soil thickness (cm), bulk density (g cm−3), TOC (g kg−1) and percentage of the >2 mm fraction, respectively. In our C model, changes of SOCD were calculated by the differences of TOC between control and transplanted samples. The SOCD distribution was displayed using histogram, and the average SOCD change and its uncertainty were estimated by commanding ‘one.boot’ in the R package simpleboot (Peng, 2008).
Vegetation variables were measured in a selected quadrat in the plot. Vegetation species, density, biomass and average height were recorded according to an established protocol (Klein et al., 2007). Plant aboveground biomass was estimated using a non-destructive sampling method (Klein et al., 2007).
DNA extraction and GeoChip analysis
DNA extraction, purification, labeling, hybridization to GeoChip 4.0, raw data processing and statistical analyses were described in recent studies (Yang et al., 2013; Chu et al., 2014; Gao et al., 2014). In brief, DNA was extracted from soil samples at the depth of 0–20 cm using FastDNA spin kit for soil (MP Biomedical, Carlsbad, CA, USA) and then precipitated and dissolved in nuclease-free water. After labeling with fluorescent dye Cy-5, 1.0 μg DNA was hybridized with GeoChip 4.0 in a MAUI hybridization station (BioMicro, Salt Lake City, UT, USA) and scanned by a NimbleGen MS200 scanner (Roche, Madison, WI, USA). GeoChip data were normalized by relative abundance, that is, each spot divided by total intensity of its microarray and then multiplied with average value of microarray’s total intensity. After that, the data were transformed by natural logarithm.
Results
The transplant effects on vegetation, soil variables and microbial community
Nearly all soil transplants showed a significant increase (P=0.006) in total aboveground vegetation biomass, total vegetation coverage and vegetation species number (Supplementary Table S1). Similarly, a significant increase in the concentration of TOC, total N and ammonia at the soil depths of 0–10 and 10–20 cm was also observed (Supplementary Table S1). A significant increase was also observed in the efflux of CO2 and N2O, whereas CH4 efflux remained unchanged. A significant positive correlation between CO2 efflux and TN_10 cm (r=0.69, P<0.001) was observed, as recorded previously (Bragazza et al., 2006).
Overall microbial functional diversity (Shannon and Simpson indices) was significantly lower in transplanted samples than control samples (P<0.003; Supplementary Table S2). Also, microbial community structures were clearly changed, with transplanted samples showing significantly different community structure compared with their in-place controls (Supplementary Figure. S1; Adonis P<0.081). In addition, changes in microbial community structure did not show linear correlations with geographic distance or changes in temperature (Supplementary Figure. S2). For example, both distance and change in temperature were less between the 3800 and 3600 m sites when compared with the 3800 and 3400 m sites, yet microbial community structure in the soil transplanted from 3800 to 3600 m showed a significantly greater change compared with soil transplanted from 3800 to 3400 m. The functional gene structure of the microbial communities was significantly (P=0.001) correlated with environmental variables, but not with geographic distance or temperature (Table 1). Interestingly, a similar non-linear response has been observed in previous soil warming experiments (Xiong et al., 2014).
The transplant effects on N cycle genes
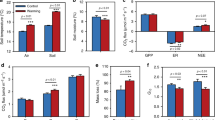
Overall, genes associated with N cycle demonstrated significantly lower relative abundance (P<0.050) in transplanted samples. However, specific genes showed differing responses, for example, the relative abundance of ureC increased, whereas gdh decreased in transplanted samples (Figure 1a). The combined effect of these two changes potentially could result in an increase in urea ammonification and hence the N mineralization, because ureC protein converts urea into ammonia, and the gdh protein converts α-ketoglutarate and ammonia to glutamate. Consistently, the ammonia content and pH were higher at warmer sites (Supplementary Table S1), thus microbe-available ammonia was increased, as microbes preferentially used NH3-N but not NH4+-N (He et al., 2012). Almost all of other N cycle genes were less abundant at warming sites except the nitrate reduction gene narG. In accordance, the NO3−-N content was lower and TN content was higher at the warming sites (Supplementary Table S1). Notably, the lower abundance of N fixation gene nifH at warming sites was consistent with a previous finding, showing that increased N suppressed N fixers (Walker et al., 2008).
(a) The warming effect on N cycle genes. The percentages in brackets indicate changes in average abundances of functional genes between warming and control sites. Red and green represent the increase and decrease in the average abundance by warming, respectively. The gray-colored genes are not targeted by GeoChip. The differences between warming and control sites were analyzed by two-tailed paired t-tests. ***P<0.001, **P<0.01, *P<0.05. (b) Correlations between N cycle gene abundances and N2O efflux. Scatterplots of N2O efflux vs abundances of gdh, ureC and amoA genes are shown along with linear regression lines.
Pearson correlation analyses showed that N2O efflux had positive correlations with ureC (P=0.001) and ammonium-oxidizing gene amoA (P=0.029) but a negative correlation with gdh (P=0.001) (Figure 1b), whereas all other N cycle genes showed no significant correlation. Thus, ammonification and subsequent nitrification appeared to contribute to N2O emission, resulting in a significant (P=0.040) increase of N2O emission at warming sites (Supplementary Table S1).
The transplant effects on C cycle genes
The total abundance of all C cycle genes decreased (Figure 2a). Among these, a number of C degradation genes were significantly (P<0.050) decreased (Figure 2b), coinciding with the increase of TOC (Supplementary Table S1). Among C degradation genes, changes in cellobiase and exoglucanase involved in cellulose degradation, mannanase involved in hemicellulose degradation and acetylglucosaminidase, endochitinase and exochitinase involved in chitin degradation were most notable, suggesting that warming differentially repressed genes for degrading recalcitrant but not labile C so as to maintain long-term soil C stability and storage. All except 2 (Clostridium saccharolyticum and Acidothermus cellulolyticus) of the 26 exoglucanase genes affected by warming were derived from Ascomycota or other fungi, suggesting that exoglucanase genes shifted predominantly from bacteria to fungi. By contrast, most genes associated with chitin degradation were derived from bacteria. No correlation was observed between CO2 efflux and C cycle genes by Mantel tests or Pearson correlation analysis using relative abundances of microbial genes, which could be interpreted as suggesting that microbial respiration was not the major contributor to soil CO2 efflux for this grassland.
The warming effect on C cycle genes. Average abundances of (a) overall C functional group, (b) C degradation, (c) C fixation and (d) CH4 cycle genes are compared between warming and control sites. Error bars represent standard error (n=18). The differences between warming and control sites were analyzed by two-tailed paired t-tests. ***P<0.001, **P<0.01, *P<0.05.
Although C fixation gene abundances remained largely unchanged by warming, there was an increase in the relative abundance of pcc encoding the Propionyl-CoA carboxylase to produce (S)-methylmalonyl-CoA through incorporation of bicarbonate radical (Figure 2c), suggesting that it was a preferred C fixation pathway under warming. Most of pcc genes affected were derived from taxa associated with Actinobacteria, Bacillus and Alpha-proteobacteria.
For CH4 cycle genes, the total abundance of mcrA for CH4 production and pmoA and mmoX for CH4 oxidization decreased (P=0.050; Figure 2d), which would have resulted in no change in net CH4 efflux (Supplementary Table S1). Almost all of mcrA genes were derived from uncultured Euryarchaeota or Archaea, whereas almost all of mmoX and pmoA genes were derived from uncultured bacteria.
The transplant effects on selected stress genes
Cold shock genes may be important for coping with the periodically cold alpine environment. Under the warming condition, the total abundance of cold shock genes (cspA, cspB, desR and desK), derived from a wide range of phyla including Actinobacteria, Firmicutes and Proteobacteria, significantly decreased (Supplementary Figure. S3), suggesting that cold stress in the alpine grassland was ameliorated by warming. In addition, stress genes responsive to P limitation (phoAB and pstABC) and N limitation (glnR) were also decreased. Accordingly, TN content was significantly greater at the warming sites (Supplementary Table S1).
Environmental variables linking to microbial communities
To identify environmental variables linking to microbial community structure, automatic forward selection and variance inflation factors were used to select variables independent of other variables, resulting in 12 variables (annual air temperature, soil temperature, soil moisture, water content, soil pH, NO3−-N, ammonia, TOC, TN, vegetation biomass, total coverage of plants and species number). Subsequently, canonical correspondence analysis was performed, resulting in a significant (P=0.005) model (Figure 3). A total of 38.6% variance by the first two axes was constrained in the canonical correspondenceanalysis, and the results were supported by Mantel test at P<0.050 (Table 2). Moisture, ammonia, TOC_10 cm, TOC_20 cm, TN_20 cm, C/N ratio, vegetation biomass and N2O appeared to be strongly linked to microbial community structure. Collectively, these results suggested that microbial community was correlated with soil C and N storage, which would support a specific influence.
Linkages between microbial community and environmental variables. Canonical correspondence analysis (CCA) displayed microbial community (symbols) and environmental variables (arrows). The value of the axis is the variance percentage explained for the axis. The significance of each environmental variable was calculated by ANOVA. **P<0.01, *P<0.05. T, temperature; TOC, total soil organic C; TN, total soil N; TCOV, total coverage of vegetation.
Modeling of topsoil C stock
To examine whether our observations were representative of Tibetan grasslands at the plateau level, we fed SOC measurements at the study site to a satellite-based SOC model (Yang et al., 2009) and examined whether it was consistent with the findings in the plateau-wide survey of topsoil C stock. On average, SOC stock in the top 20 cm depth of the four elevations in the alpine grasslands was 5.920 kg Cm−2. SOC changes for the downward transplanting experiment were estimated at 0.072 kg Cm−2, ranging from −0.100 kg Cm−2 to 0.300 kg Cm−2. Uncertainty analysis revealed SOC changes fluctuated between −0.003 kg Cm−2 and 0.104 kg Cm−2 at the 99% confidence level using basic bootstrap method (Figure 4), which validated the results of the survey that topsoil C stocks remained relatively stable but slightly increased at the plateau level.
Topsoil C stock modeling of the Tibetan grasslands based on a satellite-based approach (Yang et al., 2009). Bar and solid line represents the frequency and distribution trend of SOC change in Tibetan grasslands, respectively. The interval between dash lines represents the 99% confidence interval (CI) calculated based on basic bootstrap method.
Discussion
The effects of warming on terrestrial C and N pools of alpine grasslands remain elusive. Also, it has been documented that temperature affects microbial community structure and/or biomass in the alpine meadow (Lipson et al., 2002). Therefore, soil transplant was conducted in this study to examine topsoil C and N stocks in alpine grasslands and their linkages with microbial community. Four-years of warming in alpine grassland, as simulated by soil transplant, changed the relative abundances of microbial N and C cycle genes, which were concomitant with soil N and C content, and the identified mechanisms provided explanation for the stable but slightly increased trends in topsoil C stock at the plateau level. Temperature-dependent increases in vegetation biomass associated with tundra, grassland and forest ecosystems (Dukes et al., 2005) were also verified by the current study. In addition, we demonstrated that the soil microbial community structure was changed as a result of warming, which was consistent with previous studies (Xiong et al., 2014). However, while the majority of these studies have explored the impact of temperature on phylogenetic and taxonomic structure of microbial communities, this study has explicitly determined the influence of transplant-associated warming on the microbial functional potential, and linked this directly to the changes in C and N pools in these soils.
The total aboveground vegetation biomass, total vegetation coverage and species number increased in transplant soils (Supplementary Table S1), which had also been well documented elsewhere that warming stimulated vegetation growth in cold environments (Natali et al., 2012; Baldwin et al., 2014; DeMarco et al., 2014). As Mantel test showed that only total aboveground vegetation biomass was correlated with microbial community (Table 2), increased vegetation biomass could lead to an increase of fresh soil C input from litter, which might shift microbial community away from utilizing recalcitrant C. In addition, soil C loss through CO2 efflux was observed, as shown previously (Lin et al., 2009; Luo et al., 2009; Zhou et al., 2012). However, no significant correlation between CO2 efflux and C cycle genes was observed here, which could be interpreted as suggesting that microbial heterotrophic respiration was not the major contributor of soil CO2 efflux. Rather, it is likely that the increase of soil CO2 efflux was caused by soil autotrophic respiration, yet, we have no direct evidence to support this hypothesis. This is in direct contrast to arid/semiarid grasslands, where >50% of soil respiration was performed by heterotrophic activity (Zhou et al., 2012). The alpine grassland is typically a cold and wet climate, which substantially differs from the hot and dry climate of arid/semiarid grassland, and as both temperature and moisture can directly influence heterotrophic respiration (Xu et al., 2004), it is likely to be significantly lower in alpine soils, leading to greater net C storage.
Total C, N and ammonia contents in arid/semiarid grasslands were reduced under warming owing to microbial activity (Zhou et al., 2012), yet in the present study, ammonia and TN contents were significantly higher under warming. The differential increase of the abundance of ureC and the decrease of gdh may shuttle urea metabolism toward ammonia biosynthesis, resulting in an increased concentration of soil ammonia. However, we cannot assess the changes of genes associated with proteinases and deaminases, also important for N mineralization (Geisseler et al., 2010), as GeoChip 4.0 does not contain their probes. All of the three genes associated with chitin degradation, endochitinase, exochitinase and acetylglucosaminidase, decreased in abundance (Figure 2b), suggestive of a decreased need for microbes to mine chitin for its N demand. The increase of TN content was significantly correlated to the changing microbial community structure. Together, these results indicate that warming effects on the N cycle substantially differ between alpine and arid/semiarid grasslands. Notably, a recent work at our study site showed that biological N2-fixation by leguminous plants made a very small contribution to N cycle as its air temperatures during the growth season fell well below the optimum range of 20–35 oC for nitrogenase activity (Yang et al., 2011).
Nitrification appeared to contribute significantly to N2O efflux in the Tibetan grassland, as total abundances of amoA and the preceding step (ureC and gdh) were correlated to N2O efflux, which established a linkage between microbial metabolic potential and soil biogeochemical processes. This is supported by previous studies, suggesting that the nitrification process could be a dominant mechanism of grassland N2O emission (Klemedtsson et al., 1988; Yang et al., 2014). Although messenger RNA analysis is needed to determine whether these genes were being actively transcribed, recent studies have demonstrated that analyzing the gene abundance using DNA can provide strong correlations between N cycling genes and greenhouse gas (N2O) emissions (Morales et al., 2010; Liu et al., 2014; Zhao et al., 2014).
Using soil C model in this study, topsoil C stocks were predicted to remain relatively stable under climate warming, validating a soil C survey of the Tibetan grasslands (Yang et al., 2009). A general increase of vegetation biomass by climate warming has been predicted and observed worldwide (Elmendorf et al., 2012), and this ubiquity means that inconsistencies in soil C stock across different ecosystems may not be wholly driven by plant-derived C inputs. The microbial community must have an important role in controlling topsoil C stocks. Enhanced C sequestration through reducing microbial functional potentials in degrading C substrates, particularly recalcitrant C substrates, appeared to be involved in stabilizing topsoil C stock content in Tibetan grasslands.
Recently, initial efforts to include microbial processes in climate modeling have resulted in improved soil C projections at the global scale (Wieder et al., 2013). However, the predictions made for the Tibetan plateau substantially deviated from actual observations, emphasizing the need to develop a high-resolution model that explicitly includes microbial community structure and functional dynamics. The inclusion of microbial gene abundances for key metabolic pathways into climate models will likely significantly improve regional accuracy in C dynamic predictions. The mechanisms of C and N regulation identified here provide valuable insights for these predictions.
In conclusion, our findings unveil negative feedbacks that help maintain terrestrial C and N storages, which substantially differ from what is known for arid/semiarid grasslands. The incorporation of microbial functional genes, which has not been explicitly considered in C-climate models, will be useful to further improve the predictions of ecosystem feedbacks to climate warming.
References
Baldwin AH, Jensen K, Schönfeldt M . (2014). Warming increases plant biomass and reduces diversity across continents, latitudes, and species migration scenarios in experimental wetland communities. Glob Chang Biol 20: 835–850.
Balser TC, Firestone MK . (2005). Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73: 395–415.
Bragazza L, Freeman C, Jones T, Rydin H, Limpens J, Fenner N et al. (2006). Atmospheric nitrogen deposition promotes carbon loss from peat bogs. Proc Natl Acad Sci USA 103: 19386–19389.
Breeuwer A, Heijmans MPD, Robroek BM, Berendse F . (2010). Field simulation of global change: transplanting northern bog mesocosms southward. Ecosystems 13: 712–726.
Chu H, Wang S, Yue H, Lin Q, Hu Y, Li X et al. (2014). Contrasting soil microbial community functional structures in two major landscapes of the Tibetan alpine meadow. MicrobiologyOpen 3: 585–594.
De Frenne P, Brunet J, Shevtsova A, Kolb A, Graae BJ, Chabrerie O et al. (2011). Temperature effects on forest herbs assessed by warming and transplant experiments along a latitudinal gradient. Glob Chang Biol 17: 3240–3253.
DeMarco J, Mack MC, Bret-Harte MS, Burton M, Shaver GR . (2014). Long-term experimental warming and nutrient additions increase productivity in tall deciduous shrub tundra. Ecosphere 5: 5: art72.
Dukes JS, Chiariello NR, Cleland EE, Moore LA, Shaw MR, Thayer S et al. (2005). Responses of grassland production to single and multiple global environmental changes. PLoS Biol 3: e319.
Dunne JA, Saleska SR, Fischer ML, Harte J . (2004). Integrating experimental and gradient methods in ecological climate change research. Ecology 85: 904–916.
Elmendorf SC, Henry GH, Hollister RD, Björk RG, Bjorkman AD, Callaghan TV et al. (2012). Global assessment of experimental climate warming on tundra vegetation: heterogeneity over space and time. Ecol Lett 15: 164–175.
Falkowski PG, Fenchel T, Delong EF . (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039.
Feng X, Simpson A, Wilson K, WD D, Simpson M . (2008). Increased cuticular carbon sequestration and lignin oxidation in response to soil warming. Nat Geosci 1: 836–839.
Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q et al. (2014). GeoChip as a metagenomics tool to analyze the microbial gene diversity along an elevation gradient. Genom Data 2: 132–134.
Geisseler D, Horwath WR, Joergensen RG, Ludwig B . (2010). Pathways of nitrogen utilization by soil microorganisms–a review. Soil Biol Biochem 42: 2058–2067.
He J-Z, Hu H-W, Zhang L-M . (2012). Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55: 146–154.
Ise T, Dunn AL, Wofsy SC, Moorcroft PR . (2008). High sensitivity of peat decomposition to climate change through water-table feedback. Nat Geosci 1: 763–766.
Klein JA, Harte J, Zhao XQ . (2007). Experimental warming, not grazing, decreases rangeland quality on the Tibetan Plateau. Ecol Appl 17: 541–557.
Klemedtsson L, Svensson B, Rosswall T . (1988). Relationships between soil moisture content and nitrous oxide production during nitrification and denitrification. Biol Fert Soi 6: 106–111.
Lazzaro A, Gauer A, Zeyer J . (2011). Field-scale transplantation experiment to investigate structures of soil bacterial communities at pioneering sites. Appl Environ Microb 77: 8241–8248.
Li CQ, Tang MC . (1988). The climate change of Qinghai-xizang plateau and its neighborhood in the recent 30 years. Plateau Meteorol 1: 332–341.
Lin XW, Wang SP, Ma XZ, Xu GP, Luo CY, Li YN et al. (2009). Fluxes of CO2, CH4, and N2O in an alpine meadow affected by yak excreta on the Qinghai-Tibetan plateau during summer grazing periods. Soil Biol Biochem 41: 718–725.
Lipson D, Schadt C, Schmidt S . (2002). Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microb ecol 43: 307–314.
Liu S, Wang F, Xue K, Sun B, Zhang Y, He Z et al. (2014). The interactive effects of soil transplant into colder regions and cropping on soil microbiology and biogeochemistry. Environ Microbiol; e-pub ahead of print 15 January 2014; doi:10.1111/1462-2920.12398.
Luo Y, Sherry R, Zhou X, Wan S . (2009). Terrestrial carbon-cycle feedback to climate warming: experimental evidence on plant regulation and impacts of biofuel feedstock harvest. GCB Bioenergy 1: 62–74.
Melillo J, Steudler P, Aber J, Newkirk K, Lux H, Bowles F et al. (2002). Soil warming and carbon-cycle feedbacks to the climate system. Science 298: 2173–2176.
Morales SE, Cosart T, Holben WE . (2010). Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4: 799–808.
Natali SM, Schuur EA, Rubin RL . (2012). Increased plant productivity in Alaskan tundra as a result of experimental warming of soil and permafrost. J Ecol 100: 488–498.
Peng RD . (2008). Simpleboot: simple bootstrap routines. R package version 1: 1–3.
Ryba SA, Burgess RM . (2002). Effects of sample preparation on the measurement of organic carbon, hydrogen, nitrogen, sulfur, and oxygen concentrations in marine sediments. Chemosphere 48: 139–147.
Stevens CJ, Mountford JO, Gowing DJ, Bardgett RD . (2012). Differences in yield, Ellenberg N value, tissue chemistry and soil chemistry 15 years after the cessation of nitrogen addition. Plant Soil 357: 309–319.
Vanhala P, Karhu K, Tuomi M, BjÖRklÖF K, Fritze H, HyvÄRinen H et al. (2011). Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Glob Chang Biol 17: 538–550.
Waldrop MP, Firestone MK . (2006). Response of microbial community composition and function to soil climate change. Microb Ecol 52: 716–724.
Walker JK, Egger KN, Henry GH . (2008). Long-term experimental warming alters nitrogen-cycling communities but site factors remain the primary drivers of community structure in high arctic tundra soils. ISME J 2: 982–995.
Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJ et al. (2002). Ecological responses to recent climate change. Nature 416: 389–395.
Wieder WR, Bonan GB, Allison SD . (2013). Global soil carbon projections are improved by modelling microbial processes. Nat Clim Change 3: 909–912.
Xiong J, Sun H, Peng F, Zhang H, Xue X, Gibbons SM et al. (2014). Characterizing changes in soil bacterial community structure in response to short-term warming. FEMS Microbiol Ecol 89: 281–292.
Xu L, Baldocchi DD, Tang J . (2004). How soil moisture, rain pulses, and growth alter the response of ecosystem respiration to temperature. Global Biogeochem Cy 18: GB4002.
Yang B, Qiao N, Xu X, Ouyang H . (2011). Symbiotic nitrogen fixation by legumes in two Chinese grasslands estimated with the 15 N dilution technique. Nutr Cycl Agroecosys 91: 91–98.
Yang Y, Fang J, Smith P, Tang Y, Chen A, Ji C et al. (2009). Changes in topsoil carbon stock in the Tibetan grasslands between the 1980 s and 2004. Glob Change Biol 15: 2723–2729.
Yang Y, Wu L, Lin Q, Yuan M, Xu D, Yu H et al. (2013). Responses of the functional structure of soil microbial community to livestock grazing in the Tibetan alpine grassland. Glob Chang Biol 19: 637–648.
Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L et al. (2014). The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J 8: 430–440.
Zhang W, Parker KM, Luo Y, Wan S, Wallace LL, Hu S . (2005). Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Glob Chang Biol 11: 266–277.
Zhao L, Li Y, Xu S, Zhou H, Gu S, Yu G et al. (2006). Diurnal, seasonal and annual variation in net ecosystem CO2 exchange of an alpine shrubland on Qinghai-Tibetan plateau. Glob Chang Biol 12: 1940–1953.
Zhao M, Xue K, Wang F, Liu S, Bai S, Sun B et al. (2014). Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J 8: 2045–2055.
Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X et al. (2012). Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Clim Change 2: 106–110.
Zhuang Q, He J, Lu Y, Ji L, Xiao J, Luo T . (2010). Carbon dynamics of terrestrial ecosystems on the Tibetan Plateau during the 20th century: an analysis with a process-based biogeochemical model. Global Ecol Biogeogr 19: 649–662.
Acknowledgements
The authors wish to thank Haibei Research Station staff for sampling, Hao Yu for GeoChip assistance and the two anonymous reviewers and the editor for constructive comments and suggestion to make this manuscript greatly improved. This research was supported by grants to Yunfeng Yang from the National Key Basic Research Program of China (2013CB956601), Major Science and Technology Program for Water Pollution Control and Treatment (2013ZX07315-001-03), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15010102), National High Technology Research and Development Program of China (2012AA061401) and National Science Foundation of China (41471202), to Shiping Wang from the National Basic Research Program (2013CB956000) and National Science Foundation of China (41230750) and to Jizhong Zhou from the National Science Foundation of China (41430856). The development of GeoChip and associated pipelines used in this study was supported by the US Department of Energy (DE-SC0004601) and the US National Science Foundation (EF-1065844) to Jizhong Zhou.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
This study was conceived and led by SW, JZ and YY, HY, QL, XL and YY carried out GeoChip experiments and environmental measurements, YH, MW, XS and LW performed the analytical work, HY, SW, JG, JZ and YY wrote the manuscript. All authors discussed the results and their implications and commented on the manuscript as it progressed.
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yue, H., Wang, M., Wang, S. et al. The microbe-mediated mechanisms affecting topsoil carbon stock in Tibetan grasslands. ISME J 9, 2012–2020 (2015). https://doi.org/10.1038/ismej.2015.19
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.19
This article is cited by
-
Carbon and nitrogen cycling on the Qinghai–Tibetan Plateau
Nature Reviews Earth & Environment (2022)
-
Metabolic capabilities mute positive response to direct and indirect impacts of warming throughout the soil profile
Nature Communications (2021)
-
Long-term warming in a Mediterranean-type grassland affects soil bacterial functional potential but not bacterial taxonomic composition
npj Biofilms and Microbiomes (2021)
-
The role of biocrusts in nitrogen cycling on the tropical reef islands, South China Sea
Acta Oceanologica Sinica (2021)
-
Soil organic carbon and total nitrogen stocks in alpine ecosystems of Altun Mountain National Nature Reserve in dry China
Environmental Monitoring and Assessment (2019)